Molecular detection method for desulfovibro
A quantitative detection method and sulfate technology, which is applied to the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of the lack of development of sulfate-reducing bacteria detection methods and high detection costs, and achieve The effect of short detection time, high sensitivity and low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
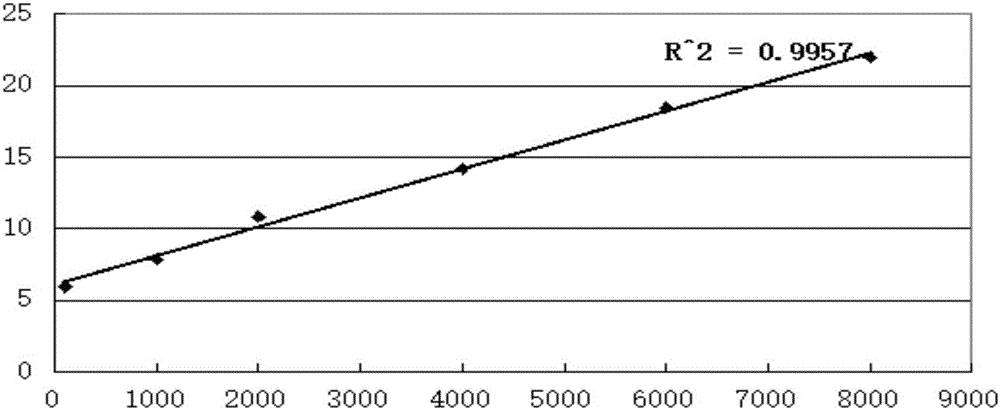
[0043] The establishment of standard curve and linear equation of the quantitative detection of embodiment 1 sulfate-reducing bacteria
[0044] 1. Test method
[0045] 1.1 Sample collection and processing
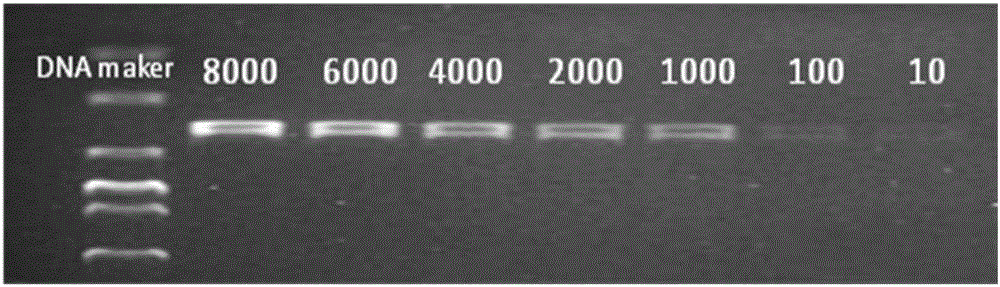
[0046] The sulfate-reducing bacteria reagent bottle was used to selectively cultivate sulfate-reducing bacteria in the oil well production fluid of a certain block of Liaohe Oilfield at 55°C. After seven days, the concentration of sulfate-reducing bacteria in the reagent bottle was detected to be 1×10 8 per ml, make the dilution gradient shown in Table 1.
[0047] Table 1 Bacterial solution dilution concentration and corresponding relative brightness value
[0048]
[0049] 1.2 Pretreatment of samples
[0050] 1. Take 1ml of the diluted bacterial solution in a 1.5ml centrifuge tube, centrifuge at 12000rpm for 10min, and discard the supernatant.
[0051] 2. Add 100 μL lysozyme to treat at 37°C for half an hour, use 20mM Tris for lysozyme, pH=8; 2mM Na 2 -EDTA buffer ...
experiment example 1D
[0062] Experimental Example 1 Determination of the Lower Limit of the DNA Template Concentration
[0063] 1. Experimental method
[0064] 1.1 Sample collection and processing
[0065] Sulphate-reducing bacteria in the oil well production fluid of a block in Liaohe Oilfield were taken, and cultured in a SRB reagent bottle at 55°C for 7 days.
[0066] 1.2 DNA extraction (for the extraction method, refer to the instruction manual of the Genome Extraction Kit of Tienensis Bacteria)
[0067] The concentration of DNA detected by NanoDrop2000 was 160ng / μL.
[0068] 1.3PCR amplification
[0069] With the extracted bacterial DNA as a template, configure the PCR amplification system according to Table 2 to amplify (ddH 2 Add after O dilution), wherein, the primer pair used is that the forward primer shown in SEQIDNo.1 and the nucleotide sequence are made up of the reverse primer shown in SEQIDNo.2 by nucleotide sequence; Wherein, Y represents base C Or T, R represents the base A or...
experiment example 2
[0078] Experimental example 2 Determination of the lower limit of the concentration of sulfate-reducing bacteria
[0079] 1. Test method
[0080] 1.1 Sample collection and processing
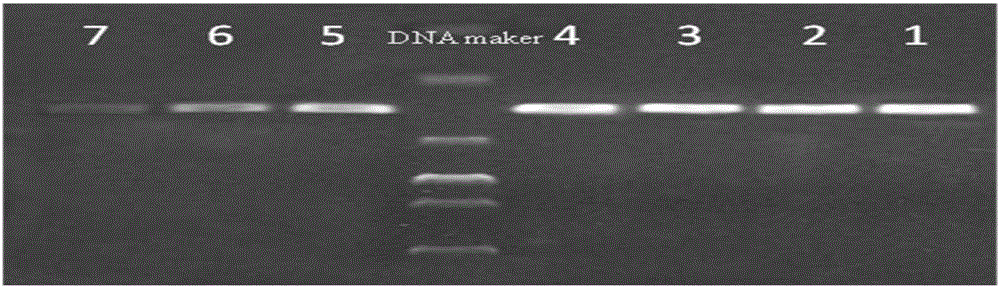
[0081] The sulfate-reducing bacteria reagent bottle was used to selectively cultivate sulfate-reducing bacteria in the oil well production fluid of a block in Karamay Oilfield, Xinjiang at 37°C. After seven days, the sulfate-reducing bacteria concentration in the reagent bottle was detected to be 2×10 5 pieces / ml. For the cultured sulfate bacteria, do the dilution shown in Table 3.
[0082] Table 3 Dilution concentration of bacterial solution
[0083]
[0084] 1.2 Pretreatment of samples
[0085] 1. Take 1ml of the diluted bacterial solution in a 1.5ml centrifuge tube, centrifuge at 12000rpm for 10min, and discard the supernatant.
[0086] 2. Add 100 μL lysozyme to treat at 37°C for half an hour, use 20mM Tris for lysozyme, pH=8; 2mM Na 2 - The EDTA buffer solution is prepared with a fina...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com