Single-stranded aptamer and application thereof
A single-stranded nucleic acid and aptamer technology, applied in the fields of biomedicine and clinical medicine, can solve the problems of decreased antibody detection ability, inability to identify irregular antibody specificity, degradation and disappearance, etc., to avoid blood transfusion reactions and prevent newborns. Hemolytic disease, the effect of avoiding destruction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
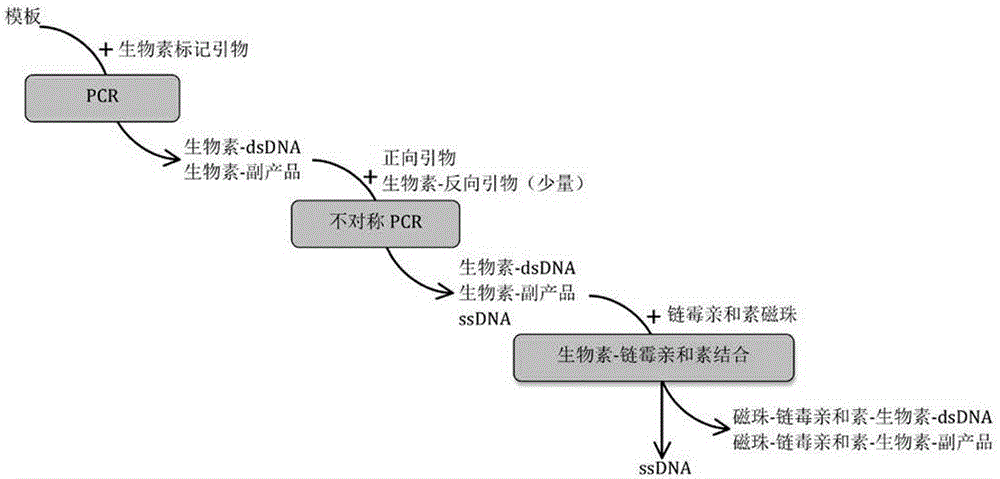
[0033] This example provides a method for preparing a ssDNA secondary library, the preparation process is shown in figure 1 , including the following steps:
[0034] Step 1: Use biotin-labeled upstream and downstream primers to perform PCR amplification on the ssDNA library to obtain biotin-carrying dsDNA to provide sufficient templates for asymmetric PCR.
[0035] The PCR reaction system is as follows:
[0036]
[0037] The Taq enzyme is selected from the kit of model M1665S of Promega Company.
[0038] The biotin-labeled upstream primer is 5'-biotin-AGAGACGGACACAGGATGAGC-3';
[0039] The biotin-labeled downstream primer is 5'-biotin-TGGATGCTGTCTTGGGGAAGG-3';
[0040] Wherein, biotin is biotin.
[0041] The ssDNA library is composed of 82 bp, the middle of the library is a random sequence of 40 bp, and the two sides are primer binding regions composed of 21 bp respectively: 5'-AGAGACGGACACAGGATGAGC-N40-CCTTCCCCAAAGACAGCATCCA-3'.
[0042] The above primers and ssDNA li...
Embodiment 2
[0079] This example provides a method for screening single-stranded nucleic acid aptamers capable of specifically neutralizing RhD antibodies through the ssDNA secondary library of Example 1: Step 1: Sequencing of the screened ssDNA sequence library
[0080] The ssDNA secondary library obtained in Example 1 was sequenced and analyzed using Ion Torrent semiconductor sequencing technology (the sequencing service was completed by Life Technologies).
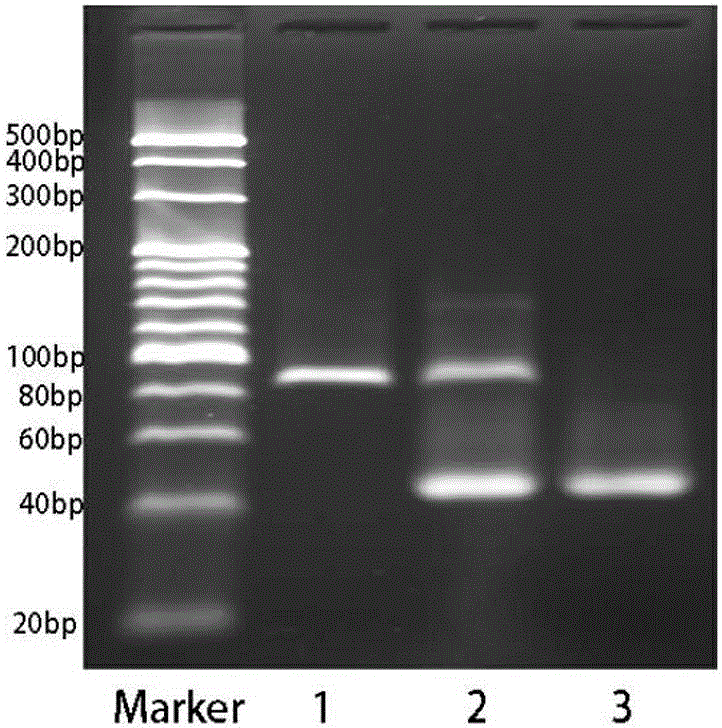
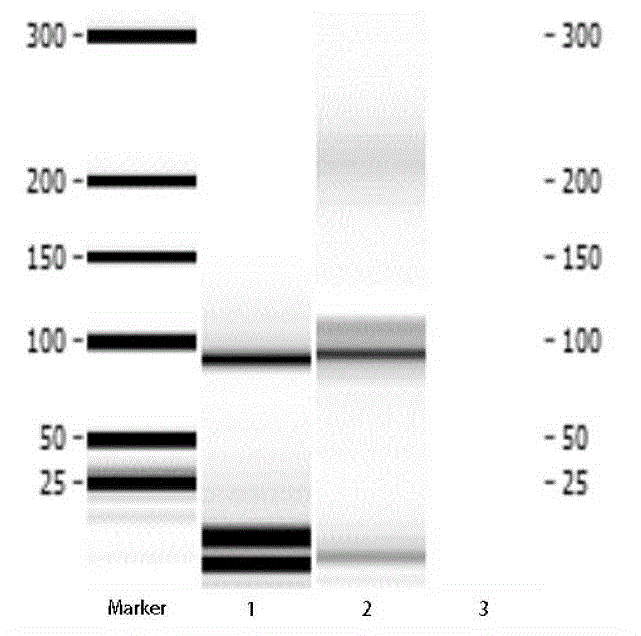
[0081] according to Figure 5 It can be seen from the ssDNA single-strand sequence library length classification statistics chart that the expected sequencing target length meets the requirements, which is about 82bp; see Figure 6 .
[0082]According to the sequencing results of the ssDNA sequence library, we took the sequence with the target sequence in the middle, adapters at both ends, and the adapters completely matched as the key verification object. At the same time, considering that the sequencing has no directionality, th...
Embodiment 3
[0156] This embodiment provides ssDNA, a kit or a chip for detecting RhD antibodies, which contain the single-stranded nucleic acid aptamer SEQ ID NO: 19 described in Example 2.
[0157] This embodiment also provides a therapeutic agent for neonatal hemolytic disease, the therapeutic agent contains the single-stranded nucleic acid aptamers SEQ ID NO: 14 and SEQ ID NO: 20 described in Example 2.
[0158] This embodiment further provides an adjuvant for blood transfusion of RhD-negative patients, the adjuvant containing the single-stranded nucleic acid aptamers SEQ ID NO: 14 and SEQ ID NO: 20 described in Example 2.
[0159]
[0160]
[0161]
[0162]
[0163]
[0164]
[0165]
[0166]
[0167]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com