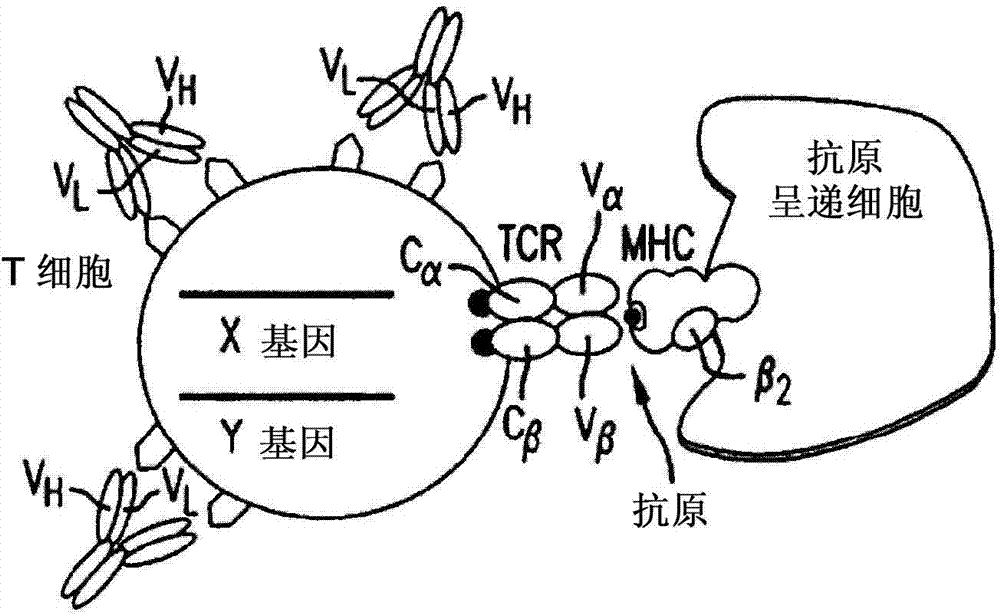
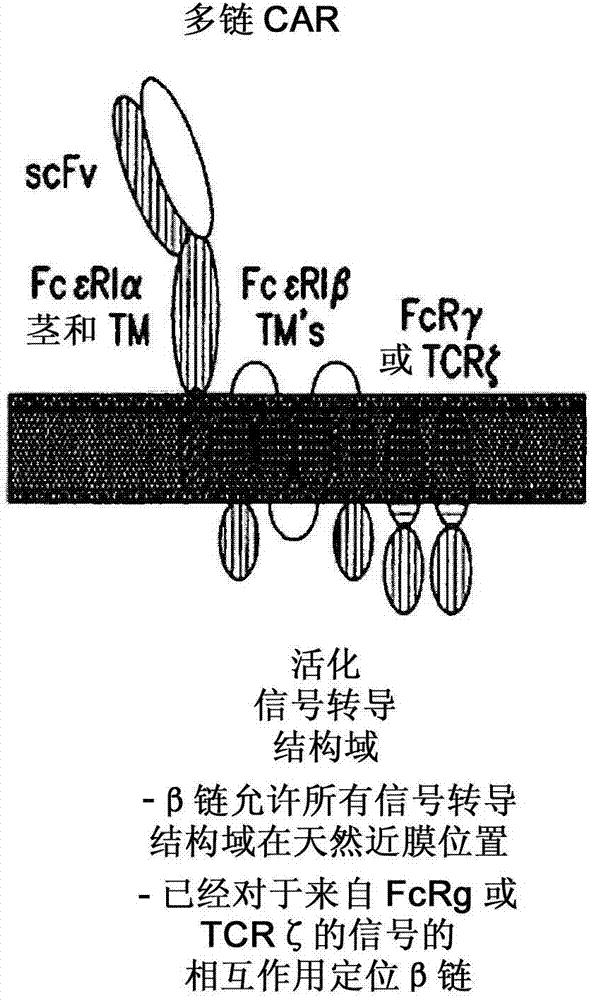
Multi-chain chimeric antigen receptor and uses thereof
A technology of chimeric antigen receptors and ligands, applied in antibody mimics/stents, anti-inflammatory agents, drug combinations, etc., can solve problems such as limiting the success of immunotherapy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0296] Embodiment 1: TALE nuclease cleaves human GR gene
[0297] Design and production of six heterodimeric TALE-nucleases targeting human GR gene exons. Table 1 below shows the target sequence cleaved by each TALE nuclease. GR TALE nucleases consist of two separate entities (termed half-TALE nucleases), each containing two 17-bp long sequences (termed half-targets) engineered to bind and cleave two 17-bp long sequences separated by a 15-bp spacer. ) consisting of repeats of the GR target sequence.
[0298]
[0299] Table 1: Description of GR TALE nucleases and TALE nuclease target site sequences in the human GR gene
[0300] Amino acid sequences and repeats of the N-terminal, C-terminal domains are based on AvrBs3 TALE (ref: GenBank: X16130.1). The C-terminal and N-terminal domains are separated by two BsmBI restriction sites. Repeat arrays (SEQ ID NOs: 7 to 18) targeting desired sequences (SEQ ID NOs: 1 to 6) were synthesized using a solid support method consisting o...
Embodiment 2
[0321] Example 2: TALE nucleases that cleave the human CD52 gene, the human T cell receptor alpha constant chain (TRAC) and the human T cell receptor beta constant chains 1 and 2 (TRBC)
[0322] Heterodimeric TALE nucleases targeting CD52, TRAC and TRBC genes, respectively, were designed and produced as described in Example 1. The target genomic sequence consists of two 17-bp long sequences (called half-targets) separated by an 11 or 15-bp spacer. Each half-target is recognized by repeats of the half-TALE nucleases listed in Table 5. The human genome includes two functional T cell receptor beta chains (TRBC1 and TRBC2). During the development of α / β T lymphocytes, one of these two constant chains is selected in each cell to splice into the variable region of TCR-β and form a functional full-length β chain. Two TRBC targets were selected in sequences conserved between TRBC1 and TRBC2 such that the corresponding TALE nucleases would cleave both TRBC1 and TRBC2.
[0323]
[0...
Embodiment 3
[0349] Example 3: TALE nucleases that cleave human CTLA4 gene and human PDCD1 gene.
[0350]Heterodimeric TALE nucleases targeting PDCD1 and CTLA4 genes, respectively, were designed and produced as described in Example 1. The target genomic sequence consists of two 17-bp long sequences (called half-targets) separated by an 11 or 15-bp spacer. Each half-target is recognized by repeats of the half-TALE nucleases listed in Table 10.
[0351]
[0352] Table 10: Description of CTLA4 and PDCD1 TALE nucleases and TALE nuclease target site sequences in human corresponding genes
[0353] Activities of CTLA4-TALE nuclease and PDCD1-TALE nuclease in HEK293 cells
[0354] Each TALE-nuclease construct was subcloned under the control of the pEF1α long promoter using restriction enzyme digestion in a mammalian expression vector. One million HEK293 cells were seeded 1 day before transfection. Two half-targets in the genomic sequence of interest in the PDCD1 and CTLA-4 genes were iden...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com