Lipid and growth trait genes
A lipid and lipophilic technology, applied in genetic engineering, plant genetic improvement, microorganisms, etc., can solve the problems of lack and difficulty in using microalgae
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0666] Example 1: Nitrogen starvation phenotype in wild-type algae.
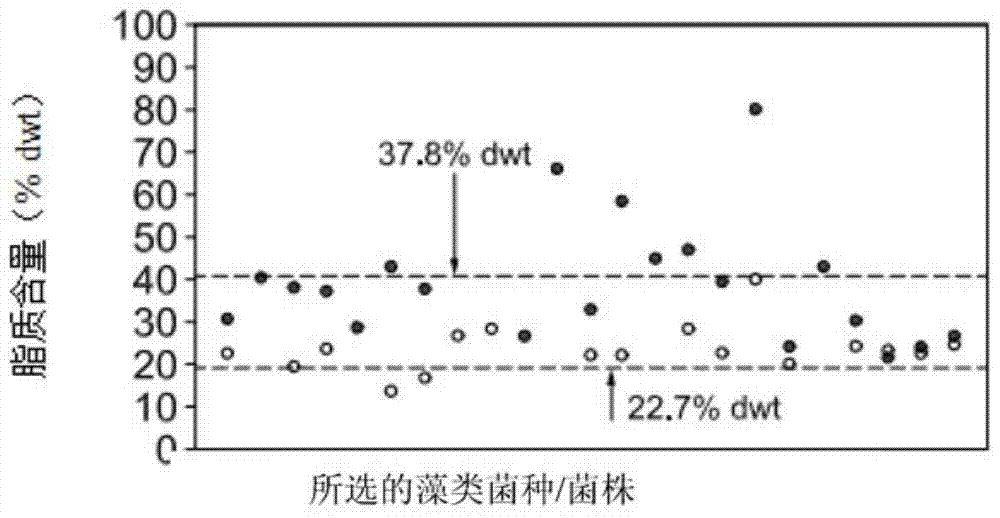
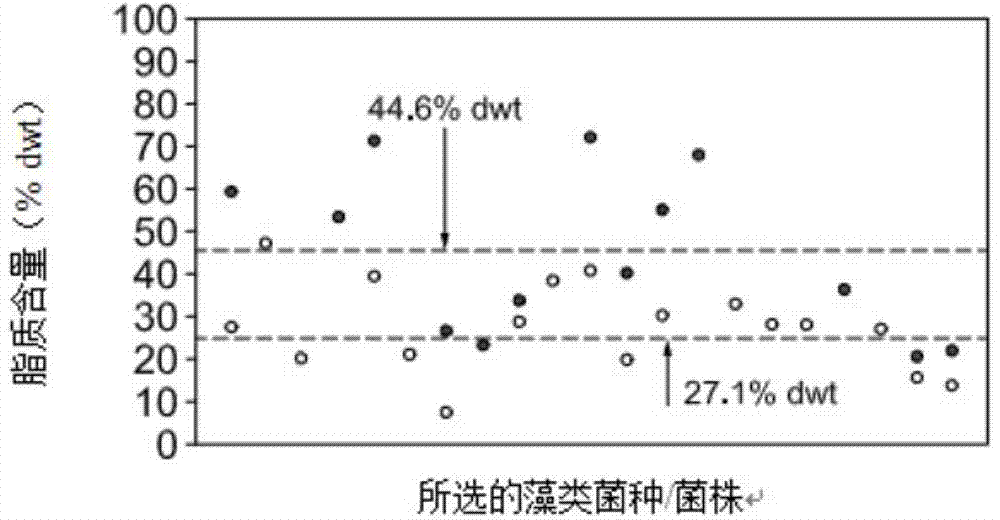
[0667] Nitrogen starvation is known to cause several phenotypes in many wild-type algal species (e.g., Dunaliella salina, Scenedesmus dimorphus, Dunaliella viridans, Chlamydomonas reinhardtii, and Nannochloropsis), including total lipid Quality increase type ( Figure 8A and 8B , Figure 41C ), reduced growth rate ( Figure 8C , Figure 41A and 41D ), and the decomposition type of chlorophyll ( Figure 8D and Figure 41B and 41E ). It is desirable to isolate these phenotypic pathways at the molecular level. For example, it is desirable to obtain an enhanced lipid phenotype without reduced growth rate and decomposed algal components.
[0668] Figure 8A Gravity fat analysis (hexane extractables) is shown. The left-hand columns of each of the two groups are in NH containing 7.5mM 4 Percent hexane-extractable lipids (%DW) after growth in medium in Cl, and in the right-hand column for each of the tw...
Embodiment 2
[0677] Example 2: Chronology of Stress Responses in Wild-Type Chlamydomonas reinhardtii at the Biochemical and Molecular Levels
[0678] In this example, the timing of biochemical and molecular reactions of wild-type Chlamydomonas reinhardtii was studied. Wild-type C. reinhardtii cells were grown in constant light in an air environment containing 5-10 L of HSM, 5% CO2, until the cells reached early logarithmic phase. The culture broth was centrifuged at 3000-5000xg for 5-10 minutes, half of the culture broth was washed with 500-1000 mL of HSM, and the other half of the culture liquid was washed with 500-1000 mL of HSM not containing nitrogen. After centrifugation again, both cultures were resuspended in a volume of medium (HSM or HSM without nitrogen) corresponding to the starting medium volume. At the time points listed in Table 2, 0.5-2 L of cells were harvested by centrifugation and analyzed for total lipid weight according to the BlighDyer method (eg BLIGH and DYER, A rap...
Embodiment 3
[0687] Embodiment 3: the method of transcriptome sequencing transcriptomics
[0688]In this example, an exemplary method for the identification of the gene encoding SN03 is described. The methods described here can be used to identify other proteins, polypeptides, or transcription factors, for example, those observed in organisms (eg, algae) that are involved in regulating or controlling different nitrogen deficiency phenotypes. These nitrogen-deficient phenotypes include, for example, increased lipid production and / or accumulation, breakdown of the photosynthetic system, reduced growth, and induction of hybridization. Genes identified as involved in the regulation or control of different phenotypes of nitrogen deficiency may have a positive or negative effect on these phenotypes, for example, increasing or decreasing lipid production, or increasing or decreasing growth rate.
[0689] In order to identify genes / proteins involved in nitrogen starvation-induced lipid phenotyp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com