Primer group and kit for carrying out spot quick detection on shrimp covert mortality nodavirus
A Nodamura virus and primer set technology, applied in microorganisms, recombinant DNA technology, microorganism-based methods, etc., can solve problems such as non-detection, and achieve the effect of less error-prone, standardized operation, and realization of programming.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
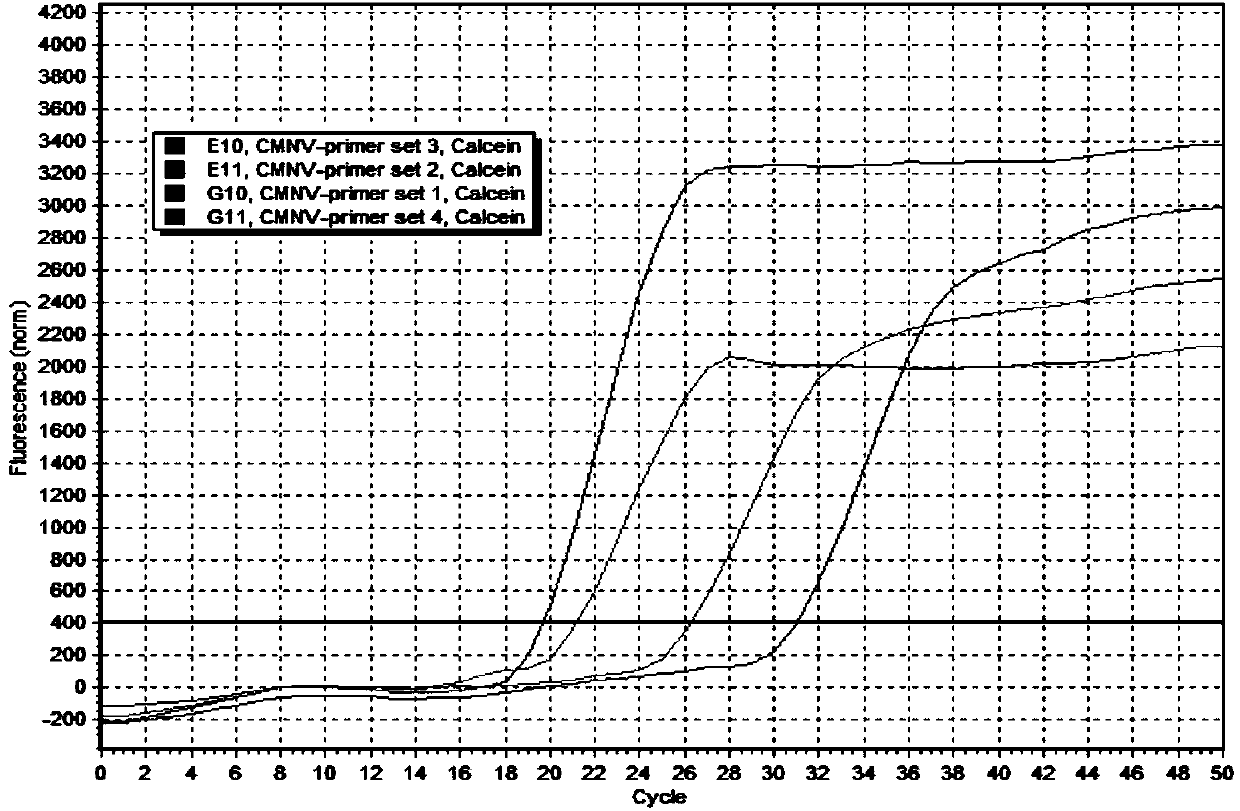
[0040] Example 1 Design and screening of on-site rapid detection primers for shrimp stealing dead wild field virus
[0041] First of all, for the CMNV protein A gene cloned from Shandong, Hebei, Fujian and other places in my country, the CMNV protein A gene was compared with the variation of the above sequence using the NCBI online program Blastn and the molecular biology software BioEitd 7.0, and the CMNV protein A gene was selected. Amplification primers were designed with the software Lamp Designer 1.02 for the conserved region sequences, and a total of 4 sets of primers were designed (Table 1).
[0042] Table 1: Amplification primers designed based on CMNV protein A gene
[0043]
[0044]
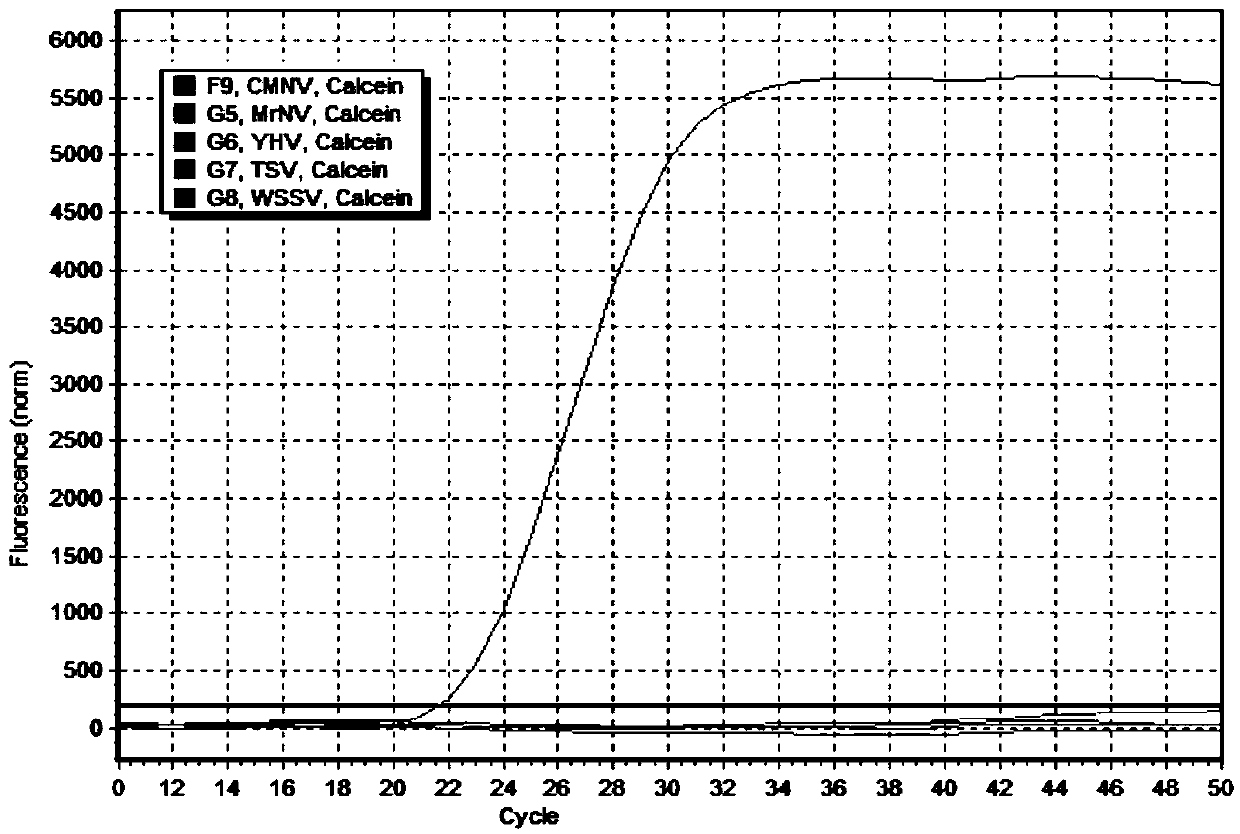
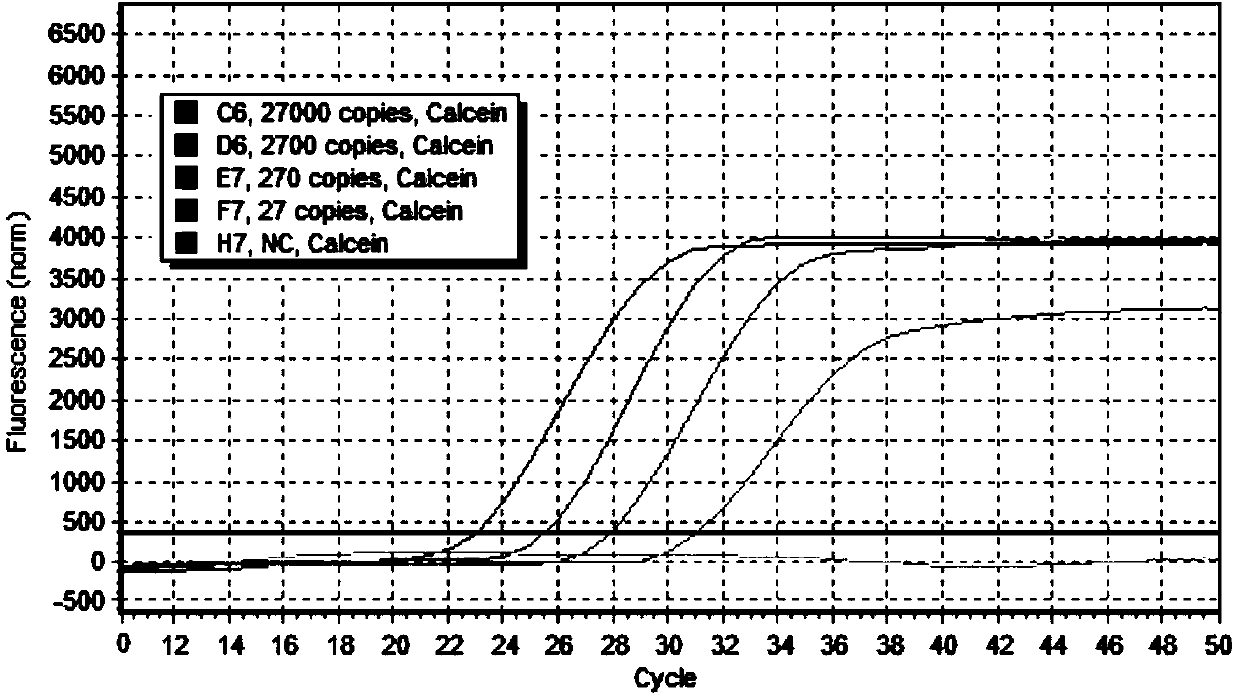
[0045] Using the 4 sets of primer combinations designed and synthesized above, the reaction system was formulated according to the following components and the amplification reaction was carried out (25 μL / reaction, referred to as method 1): each of the upstream primer 1 and downst...
Embodiment 2
[0049] Embodiment 2 Detection kit of the present invention is made up of following parts (can detect the packing of 4 samples):
[0050] (1) Sampling tubes, 4 pieces, used to hold and grind samples to be tested;
[0051] (2) Rinsing tubes, 6 pieces, each tube is equipped with 1ml of distilled water;
[0052] (3) Nucleic acid denaturation tubes, 6 pieces, each containing 20 μL of TE buffer (containing 10 mM Tris-HCl and 1 mM EDTA, pH 8.0);
[0053] (4) Amplification detection tubes, 6 pieces, each containing 24 μL of amplification reaction solution and 1 μL of dye. 0.8 μM each for melting primers 1 and 2 of the amplification primers, 0.2 μM each for upstream primer 2 and downstream primer 2 of the amplification primers, 1.4 mM each for dATP, dTTP, dGTP, and dCTP, MgCl 2 2mM, Betaine (betaine) 1.2M, Tris-HCl 20mM, KCl 10mM, MgSO 4 6mM, (NH 4 ) 2 SO 4 10mM, Triton X-100 0.1%, reverse transcriptase 5U, Bst DNA polymerase 8U; the dye is a complex formed by 50μM calcein and ma...
Embodiment 3
[0060] Embodiment 3 The detection kit of the present invention can also be made up of following parts (can detect the packing of 4 samples):
[0061] (1) Sampling tubes, 4 pieces, used to hold and grind samples to be tested;
[0062] (2) Rinsing tubes, 6 pieces, each tube is equipped with 1ml of distilled water;
[0063] (3) Nucleic acid denaturation tubes, 6 pieces, each containing 20 μL of TE buffer (containing 10 mM Tris-HCl and 1 mM EDTA, pH 8.0);
[0064] (4) Amplification detection tubes, 6 pieces, each tube is equipped with 25 μL of amplification reaction solution and 1 μL of nucleic acid dye, the components of the amplification reaction solution are as follows: each of the upstream primer 1 and downstream primer 1 of the amplification primers is 1.6 μM, Melting primers 1 and 2 of amplification primers are 0.8 μM each, amplification primers upstream primer 2 and downstream primer 2 are each 0.2 μM, dATP, dTTP, dGTP and dCTP are each 1.4 mM, MgCl 2 8mM, Betaine (betain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com