Method for detecting tomato spotted wilf virus
A technology of tomato spotted wilt virus and sequencing primers, which is applied in biochemical equipment and methods, microbe determination/inspection, DNA/RNA fragments, etc. It can solve the problems of cumbersome operation, false positive detection and false positive primers, etc., and achieve high sensitivity And the effect of high precision, easy operation and small sample volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Detect whether TSWV is contained in field samples
[0027] 1. Design specific primer sequences
[0028] Log in to the National Center for Bioinformatics (NCBI) of the United States through the Internet to query and retrieve the published N gene nucleotide sequence of TSWV, which does not contain unknown base component sequences. Use the Editseq and MegAlign software in the DNAStar 7.1 software package to edit the N gene nucleotide sequences of the selected samples, compare them one by one, and use the default parameters of the Clustal W Method (software) to perform homology on the selected N nucleotide sequences Sexual comparison to find the specific nucleotide sequence (target detection sequence) that characterizes the virus.
[0029] The design of PCR amplification primers and sequencing primers can be carried out by Assay Design SW software, and a set of primers with higher scores is used for experiments. According to the homology analysis results of DNAS...
Embodiment 2
[0046] Example 2: Detect whether TSWV is contained in imported tomato seeds
[0047] 1. Design specific primer sequences
[0048] With embodiment 1.
[0049] 2. Extract the RNA of the sample to be tested:
[0050] With embodiment 1.
[0051] 3. RT-PCR amplification with TSWV N primers
[0052] With embodiment 1.
[0053] 4. Perform agarose gel electrophoresis:
[0054] Perform agarose gel electrophoresis on the PCR product, take 2g of agarose, heat it in 100mL of electrophoresis buffer, fully dissolve, add ethidium bromide stock solution to a final concentration of 0.5μg / mL, make gel, add to the electrophoresis tank Electrophoresis buffer, so that the liquid level is just above the gel; mix 3μL ~ 6μL of PCR amplification products with an appropriate amount of sample buffer, and apply the sample; 9V / cm constant voltage electrophoresis until the bromophenol blue indicator migrates to the gel The middle part; the electrophoresis results were observed under the ultraviolet d...
Embodiment 3
[0056] Embodiment 3: Detect whether TSWV gene is contained in different virus samples
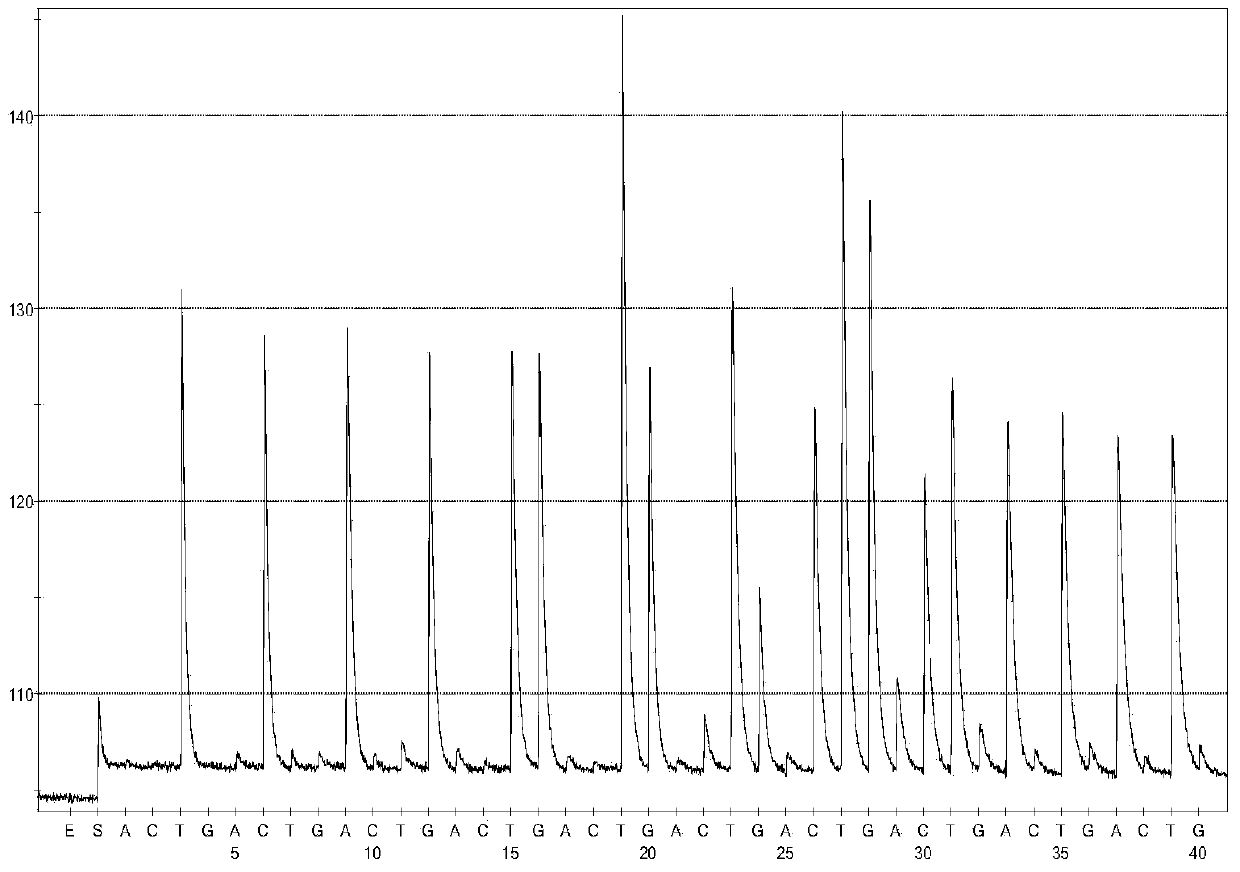
[0057] Using the pyrosequencing confirmation technology of the present invention, according to the detection steps, the tobacco ringspot virus (TRSV) isolated from imported Japanese stevia, the tomato black ring virus (TBRV) isolated from Italian cucumber seeds, and the cucumber isolated from Chilean watermelon seeds Mosaic virus (CMV) and tomato mosaic virus (ToMV), tomato yellow leaf curl virus (ToYCLV) isolated from tomato planting areas in Qingdao, and tomato spotted wilt virus-Impatiens necrotic spot virus (INSV) and Eight strains with similar genetic background to TSWV, including iris yellow spot virus (IYSV), were detected, and no amplified fragments and corresponding sequences specific to the TSWV-N gene were detected. Thereby proving that false positive results will not occur when detecting non-TSWV viruses.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com