Detection and application of a long non-coding RNA
A long-chain non-coding technology for detection reagents, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, etc., and can solve the problem of less functional research.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
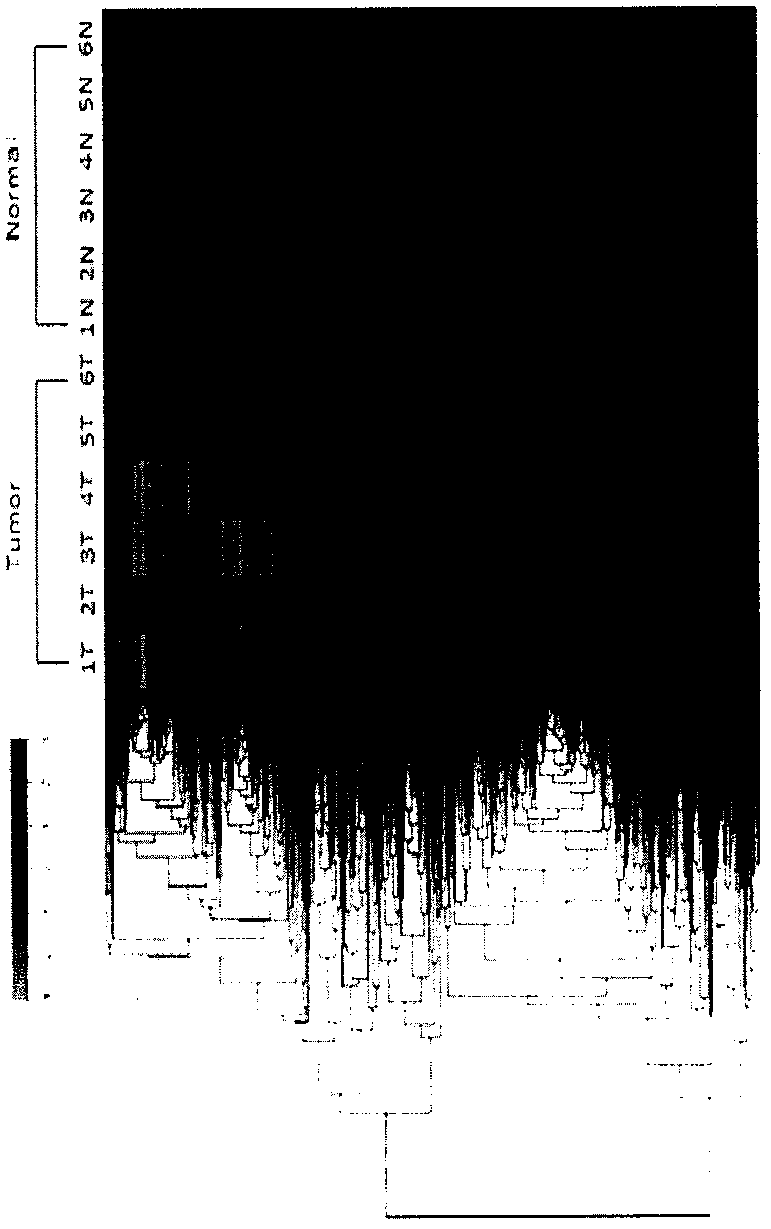
[0049] Example 1 lncRNA chip expression analysis of human esophageal squamous cell carcinoma and paired normal tissues
[0050] 1. Materials and methods
[0051] 1. Materials
[0052] Tissue samples were obtained from inpatient surgical resection samples of 6 pairs of patients with esophageal squamous cell carcinoma, each pair containing esophageal tumor tissue and paired normal esophageal tissue.
[0053] 2. Method
[0054] 2.1 Extraction of total RNA from tumor tissue and paired normal tissue
[0055] According to Qiagen's RNA extraction kit (RNeasy Micro Kit, Cat. No. 74004), the total RNA was extracted from the tumor tissues of esophageal phosphocarcinoma patients and the paired normal tissues. The kit contains RNase-Free DNase I (lyophilized), which can effectively remove the impurities of genomic DNA. The purity and concentration of the extracted RNA were quantified by NanoDrop ND-1000 Nucleic Acid Quantifier (NanoDrop Technologies, Wilmington, Delaware), and the qua...
Embodiment 2
[0078] Example 2 Preliminary verification of differential expression of HDESCC-lncRNA4 in cancer tissue and normal tissue of esophageal squamous cell carcinoma by qRT-PCR
[0079] 1. Experimental materials
[0080] Another 30 pairs (different from the samples tested by the microarray) of human esophageal squamous cell carcinoma tissues and paired normal tissues were selected, and the difference in expression of HDESCC-lncRNA4 was initially verified by qRT-PCR.
[0081] 2. Experimental methods and results
[0082] 1 Identification of primer specificity
[0083] 1.1 Screening of specific primers
[0084] (1) Extract the transcript sequence related to HDESCC-lncRNA4 from the Ensemble database, and use the primer design tool (Primer-BLAST) of NCBI to design primers according to the sequence of the transcript;
[0085] (2) The designed primers were evaluated by Oligo7, and 3 pairs of primers were designed for each;
[0086] Pair1 upstream primer: SEQ ID NO.2
[0087] Pair1 dow...
Embodiment 3
[0130] Example 3 qRT-PCR further verified the differential expression of HDESCC-lncRNA4 in cancer tissues and normal tissues of esophageal squamous cell carcinoma
[0131] 1. qRT-PCR kit composition
[0132] 1.1 Composition of dye-based HDESCC-lncRNA4qRT-PCR kit:
[0133] (1) Upstream primer: SEQ ID NO.2
[0134] (2) Downstream primer: SEQ ID NO.3;
[0135] Other reagents refer to SYBR Premix Ex Taq TM II (Tli RNaseH Plus) fluorescence quantitative kit (Code No.RR820A).
[0136] 2. Detection of HDESCC-lncRNA4qRT-PCR
[0137] 2.1 Preparation of total RNA
[0138] Select another 80 pairs of cancer tissues and paired normal tissues of patients with esophageal squamous cell carcinoma, according to the LifeTechnologies company's Reagents (Product No. 15596026) Reagents and steps required to extract total RNA, please refer to the instructions for details. The purity and concentration of the extracted RNA were quantified by NanoDrop ND-1000 Nucleic Acid Quantifier (NanoDrop ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com