GBSS1 specific enzyme activity determination method
A technology of specific enzyme activity and seeds, applied in the field of GBSS 1 specific enzyme activity determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
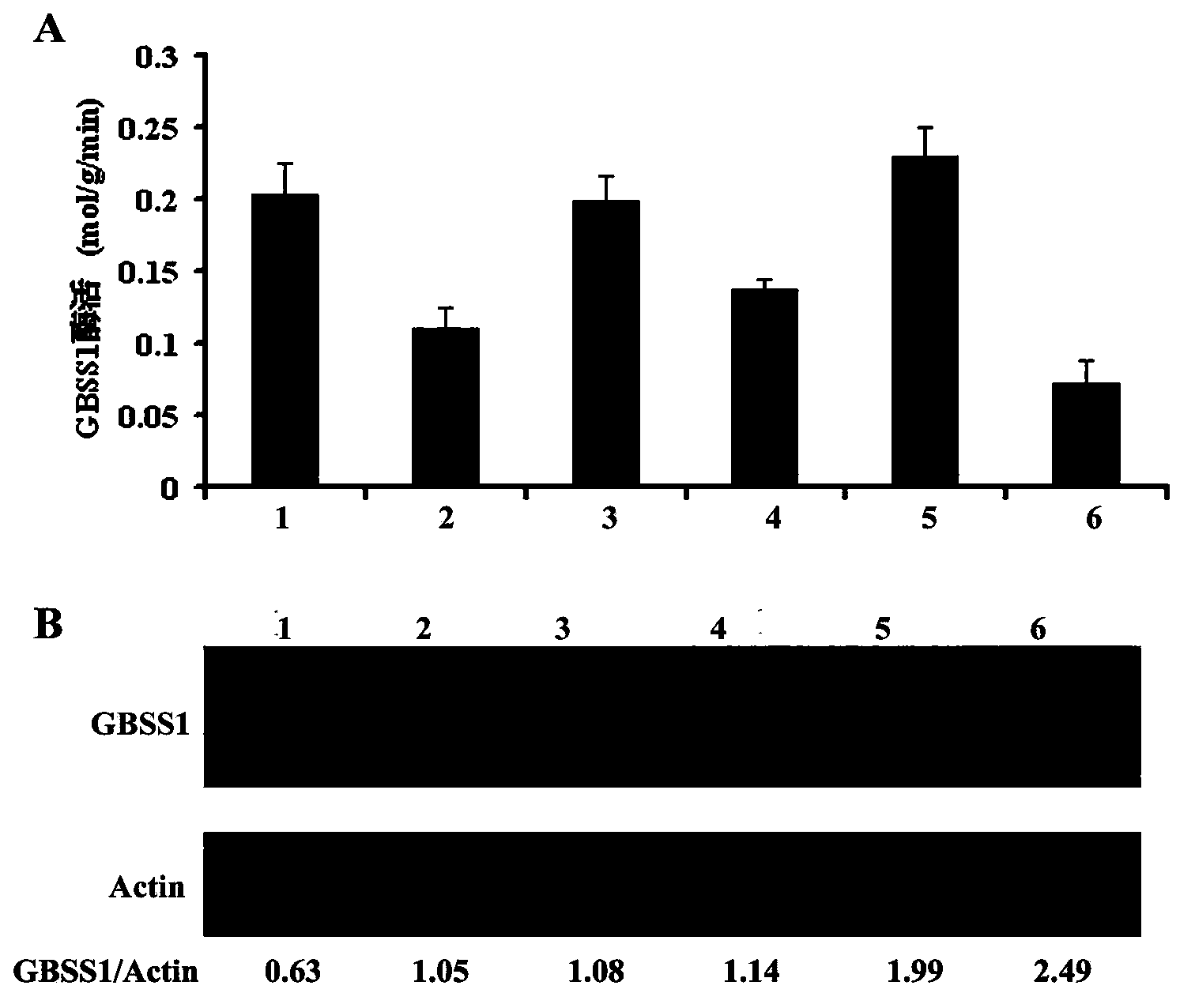
[0047] The preparation of embodiment 1, GBSS1 enzyme extract
[0048] Take 1 rice seed and freeze it in liquid nitrogen for 1 min, then manually remove the hull, seed coat and embryo, and then grind it (these steps are all carried out at 0-2°C). Three grains were taken from each rice variety as three repetitions. According to the method described by Nakamura et al. (Nakamura Y et al (1989). Plant Cell Physiol, 30:833-839), suspend the ground tissue with 1ml of extraction buffer, centrifuge at 12,000g at 4°C for 10min, and use the precipitate 1ml of extraction buffer was washed repeatedly for 2-3 times, and finally the precipitate was suspended in 100ul of extraction buffer to make GBSS1 enzyme extract.
Embodiment 2
[0049] Example 2, Quantitative Determination of GBSS1 Protein in Enzyme Extract
[0050] 2.1 Determination of total protein amount
[0051] The standard curve of bovine serum albumin was prepared according to the Bradford method (Bradford MM (1976). Anal Biochem, 72:248-254), and the rice GBSS 1 enzyme extract was diluted 1:100 to determine the amount of total protein.
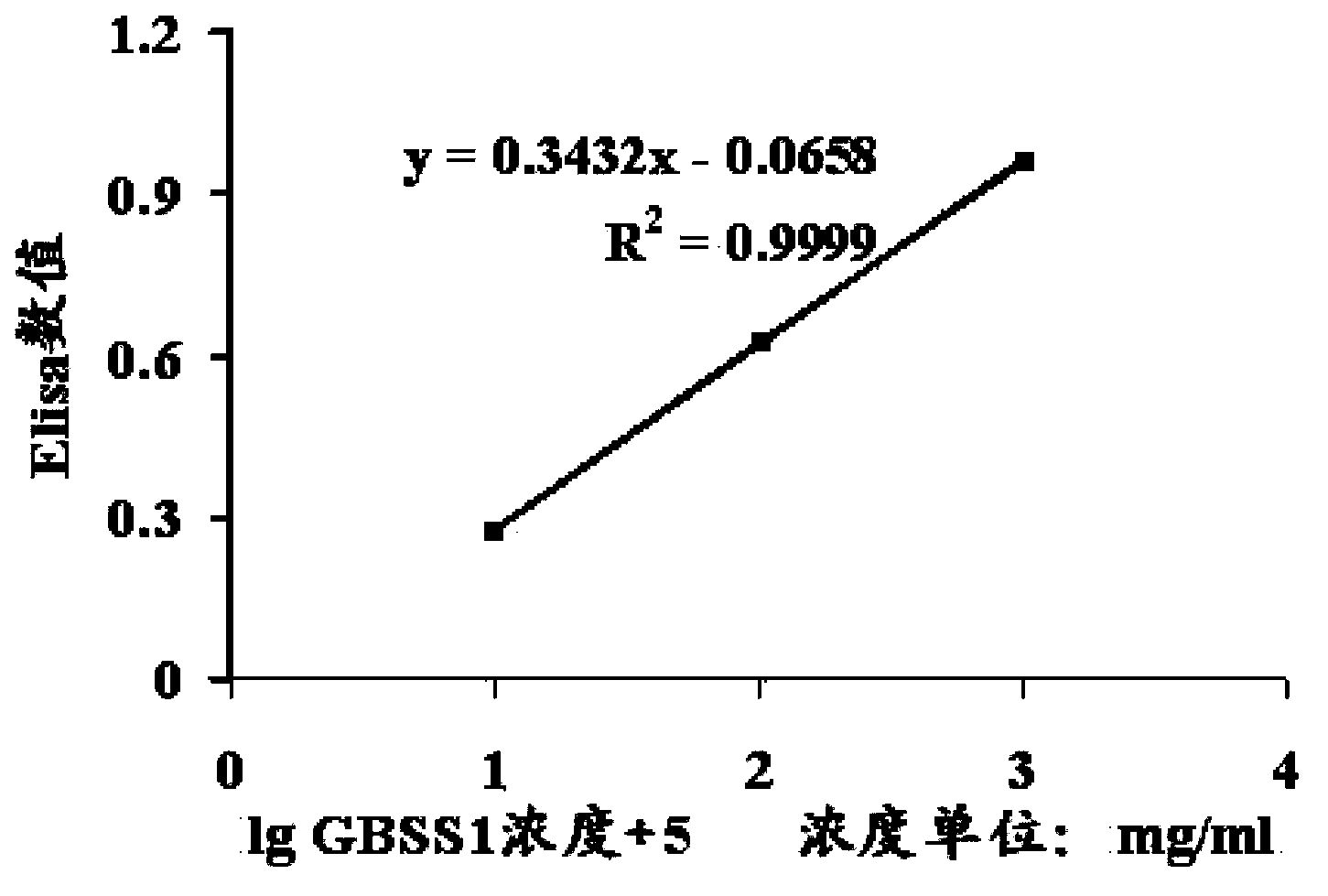
[0052] 2.2 Determination of the protein content of GBSS1 in the enzyme extract by ELISA
[0053] 2.2.1 Establishment of standard curve for prokaryotically expressed GBSS 1 protein concentration
[0054] After the prokaryotic expressed GBSS1 was purified by Ni column and Superdex200 column, the samples with GBSS1 protein content above 99% were used as standard samples. Samples were assayed for protein concentration using the Brandford method and the standard curve prepared in 2.1. Then, standard samples were serially diluted so that the protein concentration was 1.0×10 -4 -1.0×10 -2 Between mg / ml, add an e...
Embodiment 3
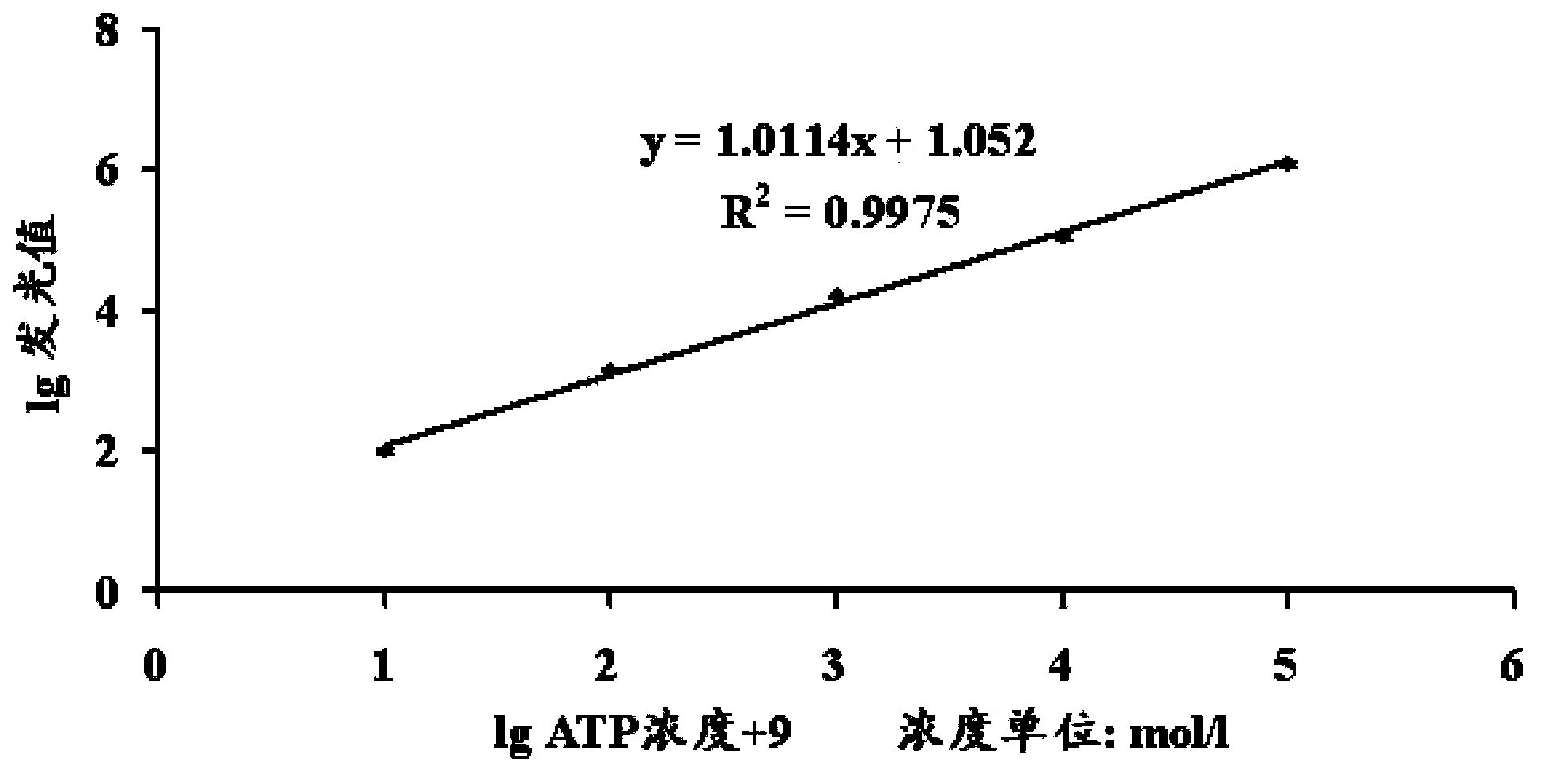
[0060] Embodiment 3, the making of ATP quantitative standard curve
[0061] Accurately weigh the ATP powder purchased from SIGMA company and prepare it into 1.0×10 -8 to 1.0×10 -4 mol / l gradient solution, and then use the SIGMA company's kit (ATP assay kit for Luciferase / Luciferin) to react, measure the luminescence value on the Promega GloMax-20 / 20Luminometer, and obtain the standard curve of the luminescence value relative to the ATP concentration accordingly . Calculation formula 3 was obtained with Excel software: lg luminescence value=1.0114×(lgATP concentration+9)+1.052.
[0062] The lg (luminescence value) / lg (ATP) standard curve that bioluminescence meter counts obtains is as follows figure 2 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com