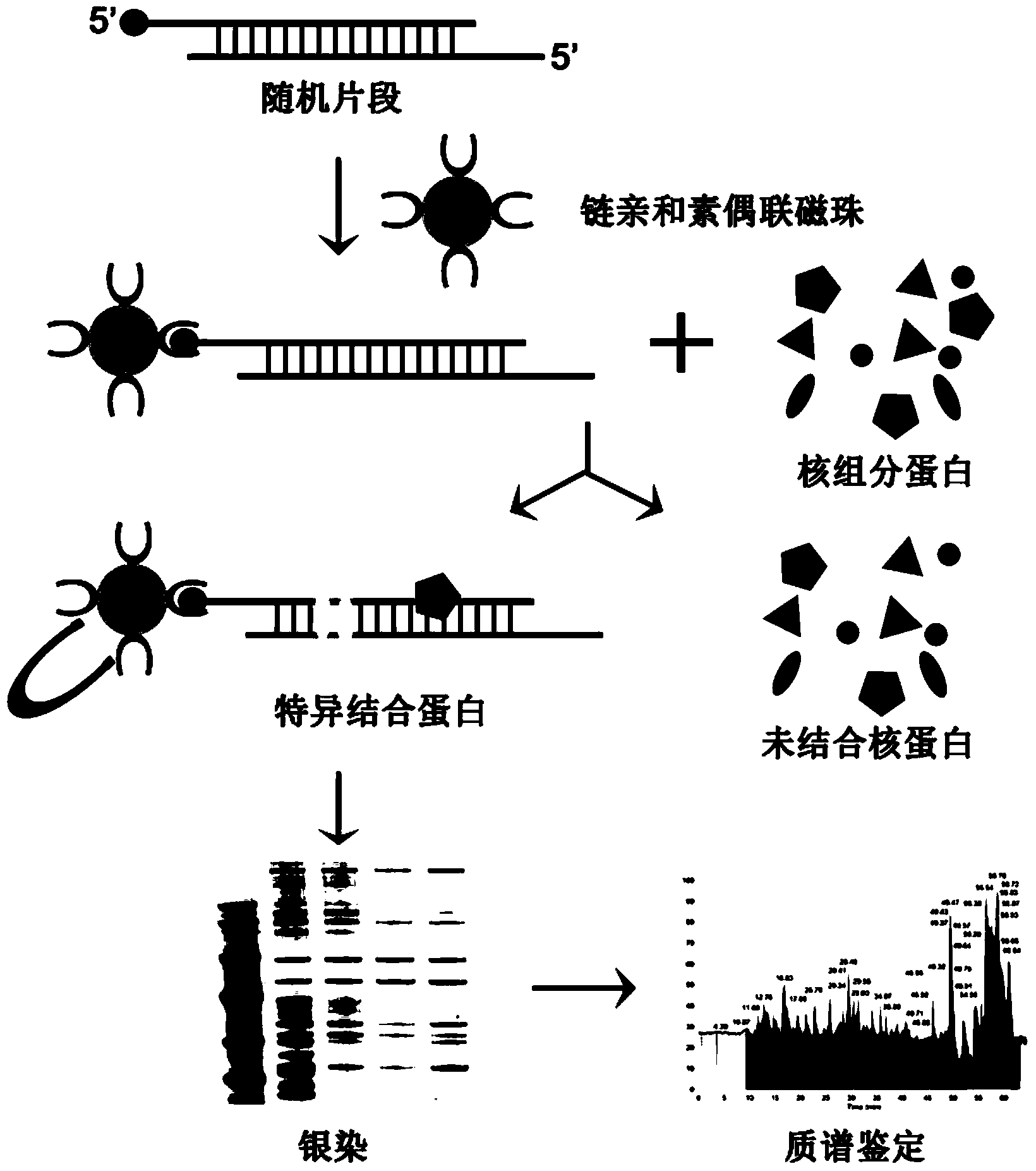
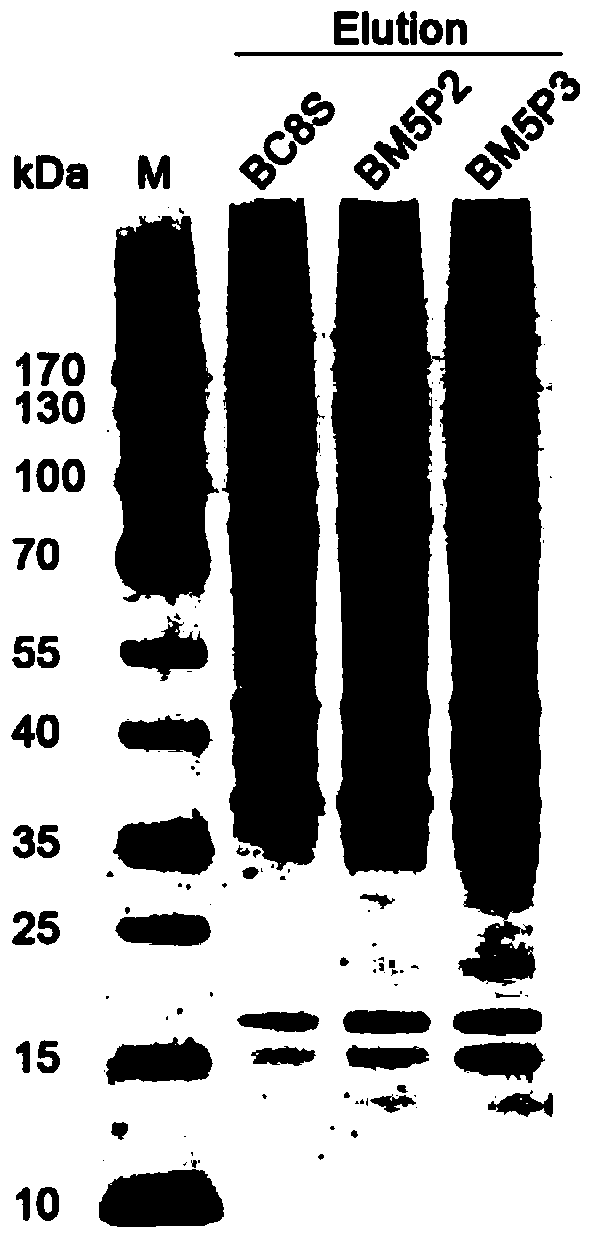Methods for separating and identifying DNA (deoxyribonucleic acid) binding protein through DNA co-immunoprecipitation
A technology for binding proteins and DNA molecules, which is applied in the field of separation and identification of DNA binding proteins to achieve the effect of improving experimental efficiency, stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
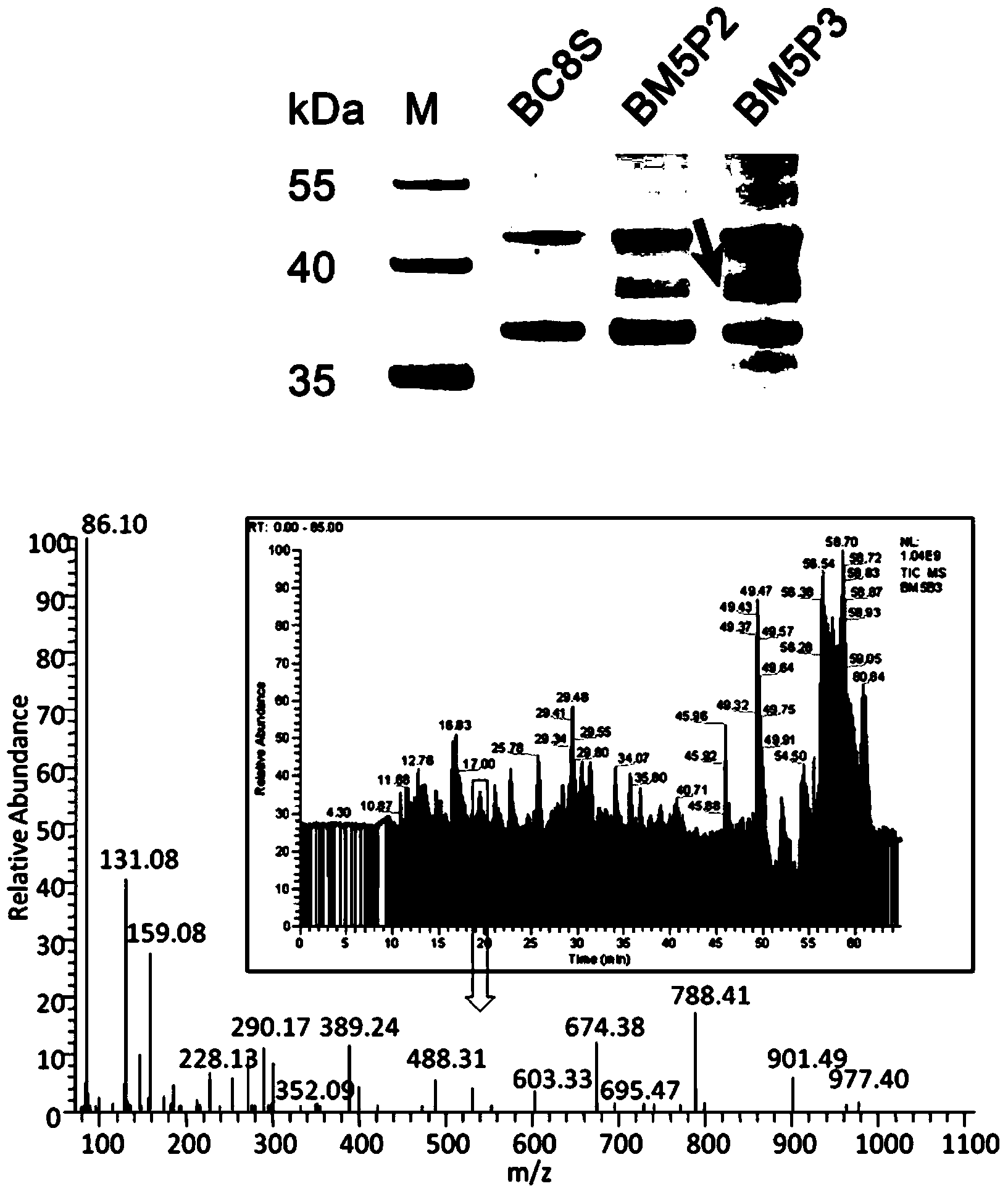
[0055] Example 1: Identification of PURA transcription factors using DNA co-immunoprecipitation combined with mass spectrometry
[0056] 1. Extraction of Nuclear Component Proteins
[0057] 1) Culture esophageal cancer cells KYSE510 (gifted by Professor Shimada of Kyoto University, Japan) in a T75 culture flask, and wash the cells with 4°C pre-cooled 1×PBS, 10ml / time×1 time. After digestion with trypsin, stop the translation of trypsin with 10ml of complete medium (RPMI1640, containing 10% fetal bovine serum), wash the cells with 10ml of PBS / time×2 times, discard the PBS and leave the precipitated cells; use 1mL Buffer A (10mM HEPES ,pH7.9,1.5mM MgCl 2 , 10mM NaCl, 0.5mM DTT) to resuspend the cells and place on ice for 15min;
[0058] 2) Add a final concentration of 0.3% NP40 lysate (50mM Tris (pH7.4), 150mM NaCl, 1% NP-40) to the cell suspension in step 1), mix well and add the cell suspension to the homogenizer , homogenate up and down 20 times;
[0059] 3) After homogen...
Embodiment 2
[0107] Example 2: Using DNA co-immunoprecipitation technology Western blotting to identify PARP transcription factors
[0108] 1. Extraction of Nuclear Component Proteins
[0109] Use a T75 culture flask to culture KYSE510 cells, wash the cells with 4°C pre-cooled 1×PBS, 10ml / time×1 time. After digestion with trypsin, stop the translation of trypsin with 10ml of complete medium, wash the cells with 10ml / time×2 times of PBS, discard the PBS and leave the precipitated cells; use 1mL BufferA (10mM HEPES, pH7.9, 1.5mM , 0.5mM DTT) to resuspend the cells and place on ice for 15min;
[0110] 1) Add NP40 lysate to the cell suspension in step 1) to a final concentration of 0.3%, mix well, add the cell suspension into a homogenizer, and homogenize up and down 20 times;
[0111] 2) After homogenization, the cell lysate suspension was reduced to 228g, centrifuged at 4°C for 10min, and the supernatant was discarded;
[0112] 3) Resuspend the nucleus pellet with 300μl Buffer B (0.3M suc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com