Alpha-amylase Amy ASS and application of alpha-amylase Amy Ass in raw starch degradation
An amylase, raw starch technology, applied in the biological field, can solve problems such as low degradation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Gene synthesis of α-amylase AmyASS
[0027] According to the whole genome information of the marine fish pathogen Aeromonas salmonicida ssp.ssp.salmonicida published in NCBI, a "putative glycoside hydrolase family protein" (amino acid sequence number YP_001143410) was found. The amino acid of the AmyASS protein is 50% similar to the experimentally identified α-amylase AmyP (Liu et al.2012), and it is speculated to be an α-amylase. The 26 amino acids predicted to be the signal peptide at the N-terminus of AmyASS protein were removed, and the gene sequence was redesigned according to the codon preference of Escherichia coli. The similarity between this gene sequence and the gene sequence (gene sequence number NC_009348.1) published in NCBI of "putative glycoside hydrolase family protein" is only 64.5%. Add NdeI and XhoI restriction enzyme cutting sites at both ends of the gene. The text sequence of the gene was sent to a biological company (Shanghai Sangon Biotechnology...
Embodiment 2
[0029] Recombinant expression of α-amylase AmyASS inclusion body
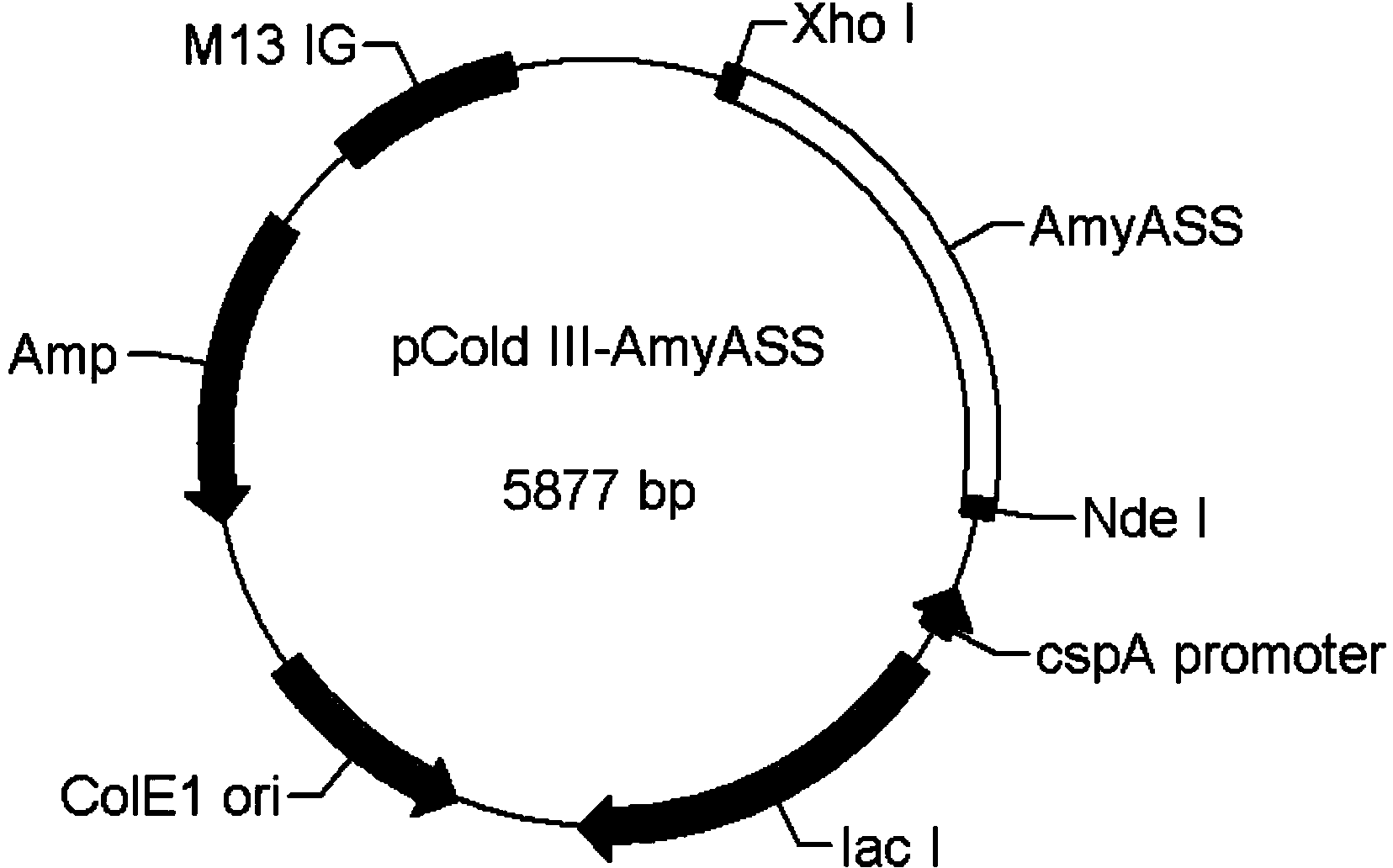
[0030] The pUC57 vector containing the α-amylase AmyASS gene and the cold shock expression vector pColdⅢ were cut with NdeI and XhoI sites respectively, and the AmyASS gene was connected to pCold In vivo, obtain the recombinant expression plasmid pColdIII-AmyASS, such as figure 1shown. The plasmid pColdIII-AmyASS was transformed into Escherichia coli BL21 cells and cultured in LB liquid medium containing 50ug / ml ampicillin at 200r / min at 37°C. When the OD600 of the Escherichia coli culture solution reached about 0.5, the culture solution was rapidly cooled to 16° C., and 1 mM IPTG was added, and then cultured overnight at 16° C. and 120 rpm. During this incubation the alpha-amylase AmyASS will be produced in the form of inclusion bodies. Collect Escherichia coli cells cultured overnight at 16°C by centrifugation at 8000r / min for 5 minutes, resuspend Escherichia coli in 50mM Tris-HCl buffer (pH8.0), and ultr...
Embodiment 3
[0033] Refolding of α-amylase AmyASS Inclusion Body
[0034] Inclusion body renaturation refers to the conversion of proteins in the form of biologically inactive inclusion bodies into biologically active proteins. The α-amylase AmyASS inclusion body was washed twice with washing solution (50mM Tris-HCl buffer, pH8.0, 5mM EDTA, 1% Triton X-100), washed once with deionized water, and the inclusion body was dissolved with lysing solution. It is required that the protein concentration after the inclusion body dissolves is about 300 μg / ml, and a certain amount of lysate is added according to the protein concentration requirement. The composition of the solution is 6M GdmCl, 100mM Tris-HCl buffer, pH6.0, pH6.0, 100mM NaCl, 100mM dithiothreitol, 1mM EDTA. Then it was placed at 4°C for 24 hours, during which it was shaken every 3 hours to promote the dissolution of inclusion bodies. After dissolving, centrifuge at 12000r / min for 20 minutes to remove a small amount of inclusion body...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com