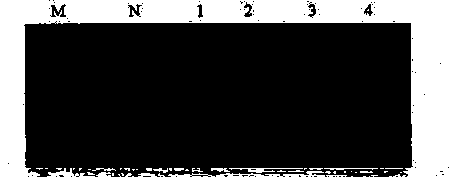Traditional Chinese medicinal material genome DNA extraction method suitable for DNA amplification technology
An extraction method and a technology of traditional Chinese medicinal materials, which are applied in the field of molecular biology, can solve problems such as cumbersome procedures, damage to the health of operators, and complicated operations, and achieve the effects of strong versatility, saving reagents and consumables, and little pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 2
[0033] 1. Materials
[0034] The materials are black snake, money snake, earth dragon, and scorpion; the method is the same as the implementation case 1, and the difference between this implementation case and the implementation case 1 is the use of the black snake specific identification primer WSS-1: 5'-GCGAAAGCTCGACCTAGCAAGGGGACCACA- 3', and WSS-2: 5'-CAGGCTCCTCTAGGTTGTTATGGGGTACCG-3'; PCR reaction system: 25 μL PCR reaction system containing 2.5 μL 10×ExTaq buffer, 2 μL 0.25 mmol / L dNTPs; 0.25 pmol each of upstream primer and downstream primer; 0.8 U Ex taq DNA polymerase, 1 μL of the above DNA template. The reaction was amplified on the 9700 gene amplification instrument of ABI company, and the cycle parameters were pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, extension at 63°C for 30 s, 38 cycles; extension at 72°C for 7 min; storage at 4°C after the reaction was completed. .
[0035] 2. Results
[0036] According to the method used in the presen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com