Mutational analysis
A technology of alleles and probes, applied in the field of analysis of genetic mutations, can solve problems such as no teaching
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
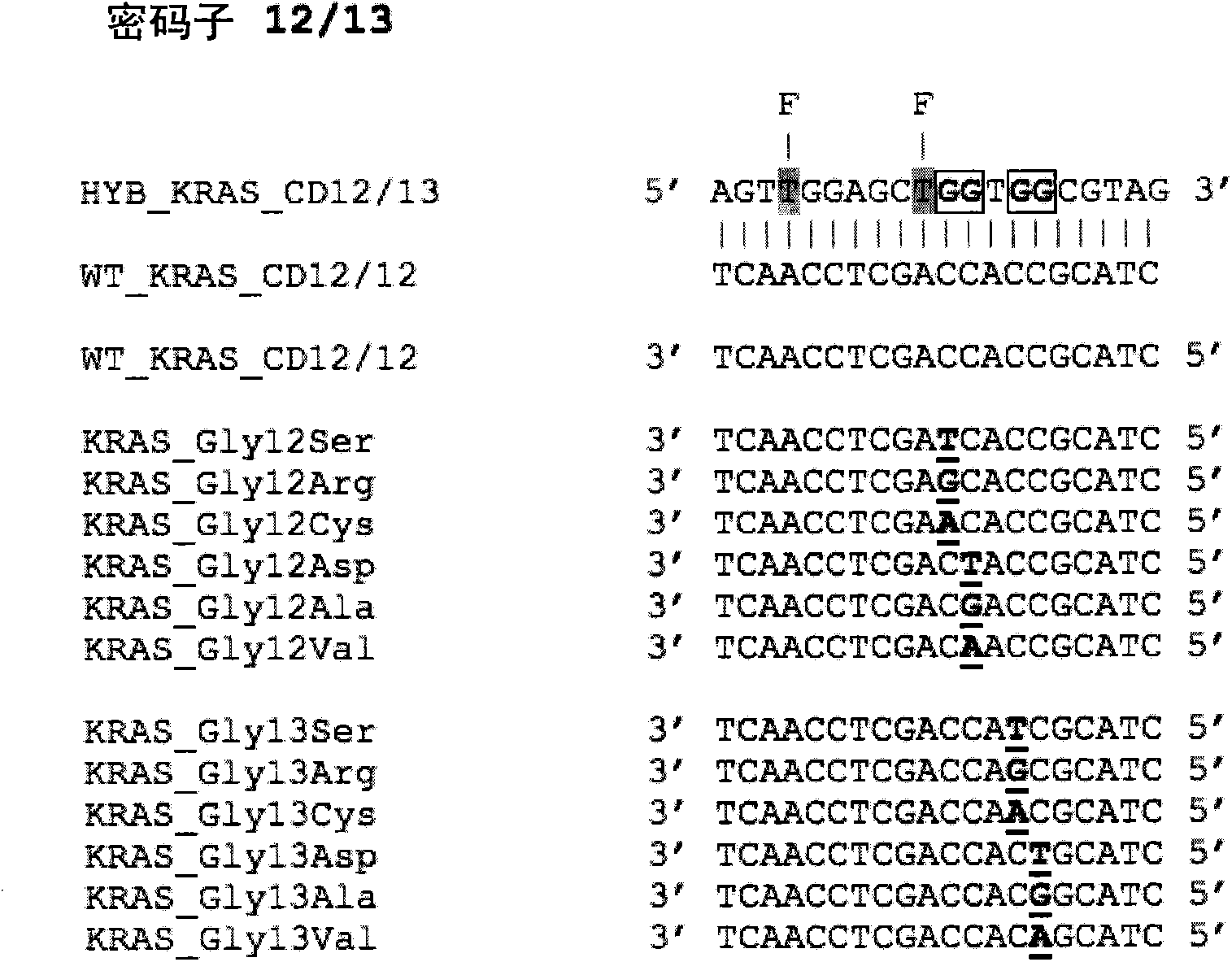
[0054] There is a need to define clinical samples to determine the direction of targeted therapy, for example, for the purpose of personalized medicine. This requires rapid and reliable screening for somatic mutations. We show here that the method can be applied to three key somatic mutations in the genes K-RAS, EGFR and BRAF. The method described in this paper utilizes Probes (which provide differential reporter signals depending on whether the probe is single-stranded or double-stranded) and asymmetric PCR (to preferentially amplify one strand of the target sequence). Typical sensitivities of the method are 1-5 copies of the mutant allele, and a SNP:wild-type ratio of greater than 5%. A single assay using a single primer set and probe can detect multiple SNPs within the same probe sequence, for example, a single probe HYB_KRAS_CD12 / 13 detects all 12 mutations in K-RAS codons 12 and 13. Example 1: K-RAS
[0055] figure 1 A partial sequence of the wild-type K-RAS gene is...
Embodiment 2-E
[0062] Example 2 - EGFR
[0063] Figure 10 The EGFR probe sequence (EGFRX18_HYB) for the EGFR exon 18 region is shown. The probe sequence is fully complementary to the wild-type region, whereas there are three possible SNP mutations (2155G>A, 2155G>T and 2156G>C), each of which differs from the probe sequence by a single mismatched base base. Likewise, the EGFRX18_HYB probe is a probe.
[0064] Figure 11 The EGFR probe sequence for EGFR exon 19 is shown (EGFRX19_HYB). Unlike exon 18, where the mutant form is a SNP, exon 19 can carry multiple deletion mutants. Possible missing regions are in Figure 11 in bold, while Figure 12 Exon 19 probes hybridizing to deletion mutants of various lengths are shown. The underlined region of this probe does not hybridize to the target and thus forms a circle. This changes the Tm of the probe:target duplex in much the same way as the presence of a base mismatch in a SNP. The change in Tm depends on the size of the unhybridized r...
Embodiment -B
[0069] Example - BRAF
[0070] The BRAF gene contained multiple potential SNP mutations at amino acid 600: 1799T>A, G or C, and multiple mutations: 1799TG>AT, 1798GT>AA or AG and 1797AGT>GAG. The probe BRAFV600_HYB is fully complementary to a part of the wild-type sequence. The probes and various target sequences are shown in Figure 17 middle.
[0071] Figure 18 The positions of the primer sequences used to amplify the BRAFV600 region are shown; these are outside the probe region. Primer targets are underlined and probe targets are in bold. The figure shows the genome sequence; the probe will be complementary, as will one of the primers.
[0072] These primers were used to amplify various samples, and real-time PCR was used to monitor the hybridization of the probes to the amplified sequences. Melting curves are shown in Figure 19 middle. All samples had a major peak at approximately 52°C, representing the wild-type sequence. The mutant sequence clusters a peak ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com