Method for distinguishing cancer cells through surface enhanced Raman spectroscopy
A surface-enhanced Raman, cancer cell technology, applied in the field of biomedicine, can solve the problems of scintillation, SERS spectral type peak position, peak intensity and peak number fluctuations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
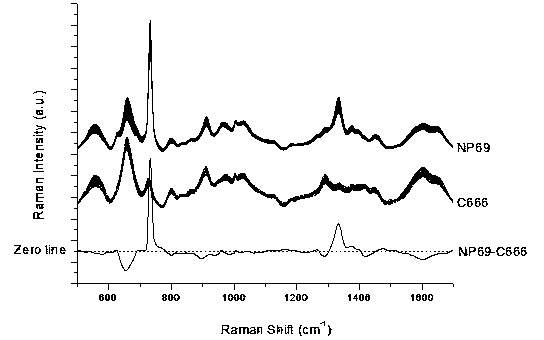
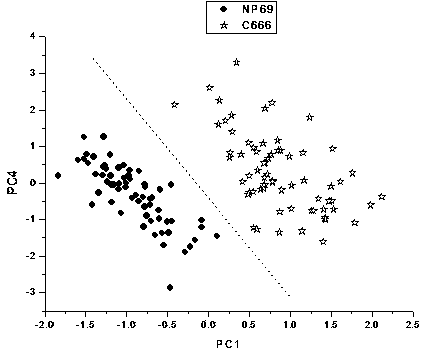
[0043] Distinguishing between normal nasopharyngeal NP69 cells and nasopharyngeal carcinoma C666 cells by surface-enhanced Raman spectroscopy
[0044] Take 100 μl and 400 μl of silver sol and normal nasopharyngeal NP69 cells or nasopharyngeal carcinoma C666 cells, mix them at room temperature, and transfer them into an electric shock cup. The electrode gap of the electric shock cup is 1 mm, and the number of cells is about 4000. Place the electric shock cup in ice bath for 5 minutes, ultrasonic pretreatment for 30 seconds, then place it in the electroporator, select manual control, apply a voltage of 350V, and last for 100ms. After electroporation, use 400μl RPMI 1640 cell culture medium to wash out the cells immediately After placing the sample pool in an ice bath for 5 minutes, transfer it to a preheated incubator, incubate at 37°C for 10 minutes, and then use a confocal Raman spectrometer for single-cell surface-enhanced Raman spectroscopy detection. The wavelength of the l...
Embodiment 2
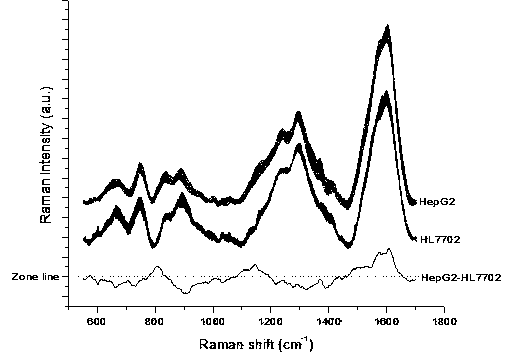
[0046] Differentiation of normal liver HL7702 cells and liver cancer HepG2 cells by surface-enhanced Raman spectroscopy
[0047] Take gold sol, 250 μl and 650 μl of normal liver HL7702 cells and liver cancer HepG2 cells, mix them at room temperature, and transfer them into an electric shock cup. The electrode gap of the electric shock cup is 4 mm, and the number of cells is about 6000. Place the electric shock cup in ice bath for 8 minutes, ultrasonic pretreatment for 60 seconds, then place in the electroporation instrument, select manual control, apply a voltage of 880V, and last for 50 ms. After electroporation, wash the cells with 800 μl RPMI 1640 cell culture medium immediately After taking out the sample pool, put it in an ice bath for 8 minutes, transfer it to a preheated incubator, and incubate at 37°C for 15 minutes, and then use a confocal Raman spectrometer to perform single-cell surface-enhanced Raman spectroscopy detection. The wavelength of the laser used to detec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com