Kit for detecting common mutations of CYP4V2 gene
A technology of kits and reagents, applied in the field of kits for detecting common mutations of the BCD pathogenic gene CYP4V2 gene, can solve problems such as lack of
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Genomic DNA Extraction
[0050] Genomic DNA in venous blood was extracted using a spin-column blood genomic DNA extraction kit (TIANamp Blood DNA Kit) produced by Beijing Tiangen Company. The reagent composition is shown in Table 2.
[0051] Table 2 Kit Components
[0052] Reagent name
[0053] Rinsing solution PW
[0054] The steps of extracting whole genome DNA in venous blood are as follows:
[0055] 1. Take 200ul of blood sample into a 1.5ml sterile centrifuge tube, add 20ul of proteinase K solution, and mix well.
[0056] 2. Add 200ul buffer solution GB, fully invert and mix well, bathe in 56 degree water for 10min, during which invert several times.
[0057] 3. Add 200ul of absolute ethanol and mix thoroughly by inversion.
[0058] 4. Put the solution obtained in the previous step into the adsorption column CB3 (put CB3 into the collection tube first), centrifuge at 12000 rpm for 30 sec, discard the waste liquid, and put CB3 into ...
Embodiment 2
[0068] Example 2 PCR amplification target gene fragment
[0069] 1. Primer sequence
[0070] Specific PCR amplification primers were designed according to the target gene fragment, and the primer sequences are shown in Table 4.
[0071] Table 4 PCR primer sequences
[0072] Primer name
Primer sequence
CYP4V2-7 F1
AGCCTATGTTGTCGAAATGT
CYP4V2-7 F2
AAAAGCAAGTCAAAGAAAGGC
CYP4V2-8 F1
CACAGTGCAGTCATCAAATC
CYP4V2-8 F2
CCAAACATACCAAACACGTC
CYP4V2-9 F1
CTTTTTAGATGTCTGCACCC
CYP4V2-9 F2
AGCATACTTACCCCACTTCAC
[0073] Note: F1 is the forward primer; F2 is the reverse primer. See NG_007965.1 for the DNA sequence data of the CYP4V2 gene.
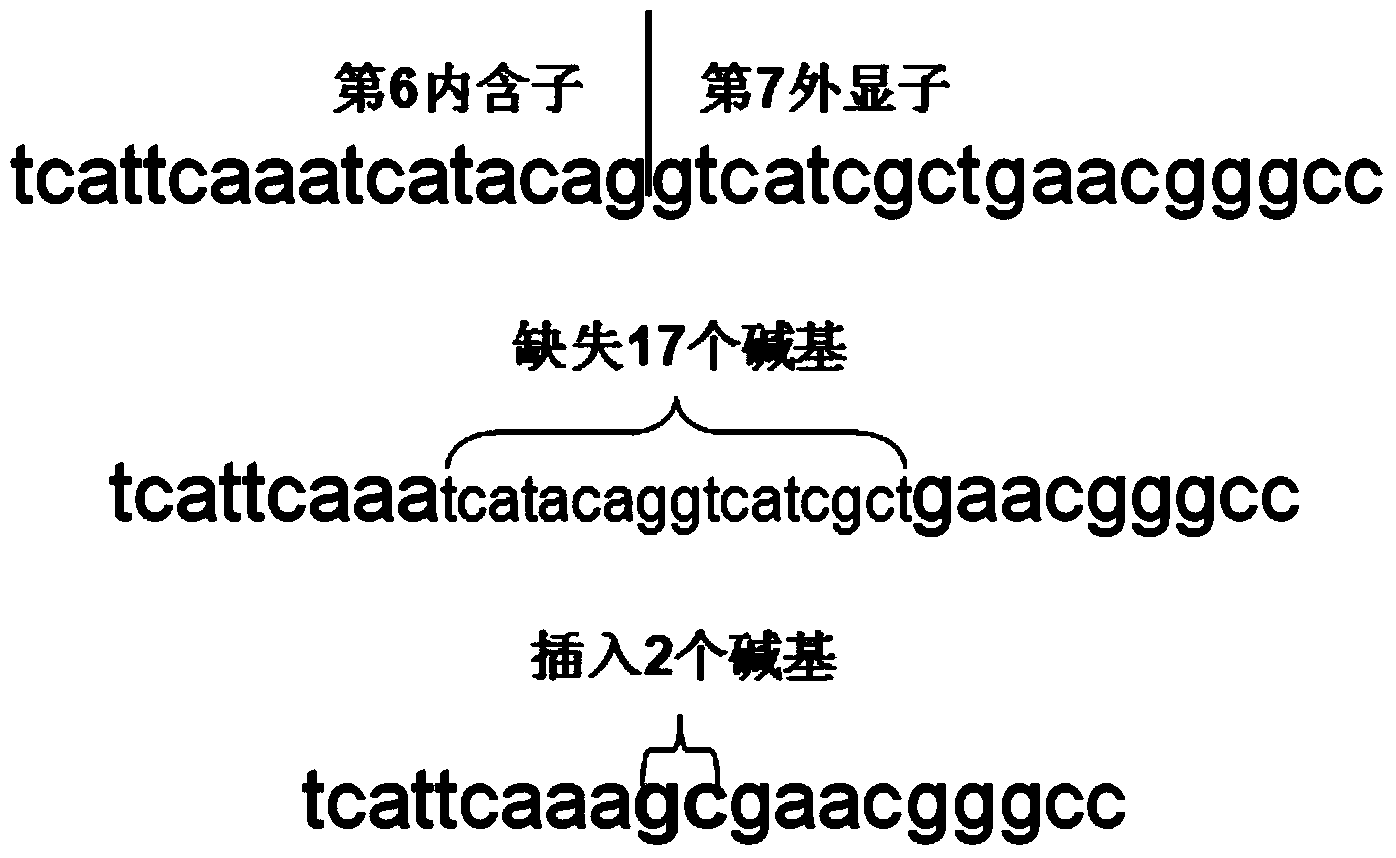
[0074] The primer pair CYP4V2-7 F1 and CYP4V2-7 F2 correspond to the detection of the mutation c.802-8_810del17insGC, and the product is the CYP4V2-7 gene with a size of 246bp; the primer pair CYP4V2-8 F1 and CYP4V2-8 F2 correspond to the detection of the mutation c.9...
Embodiment 3
[0091] Restriction enzyme digestion reaction of embodiment 3PCR reaction product
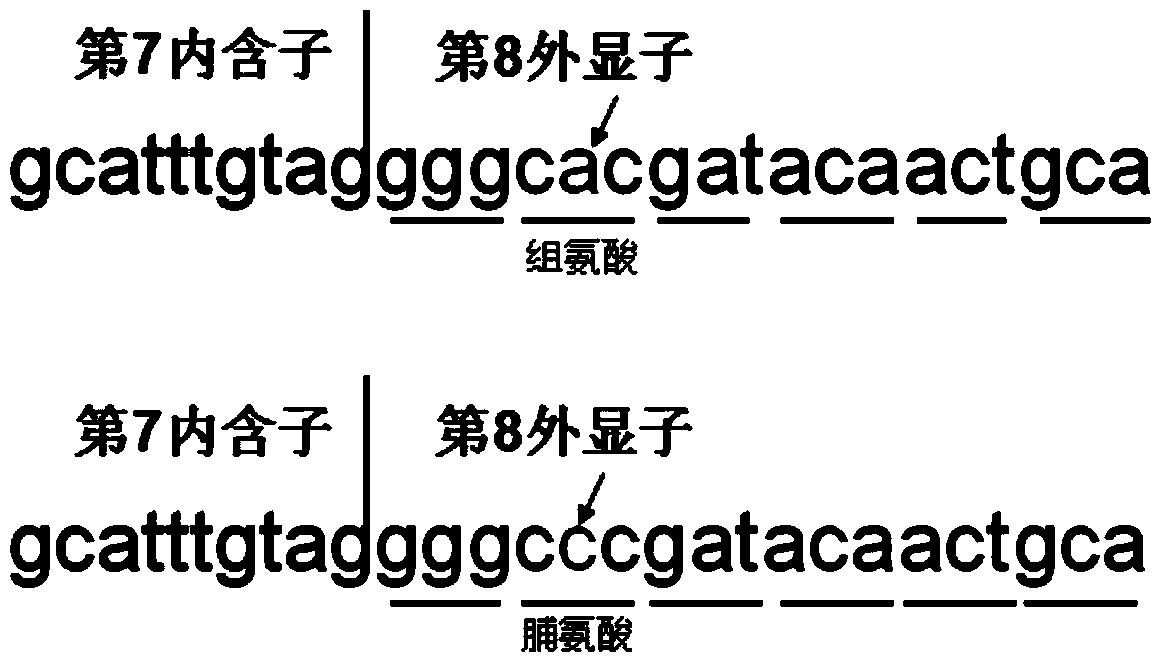
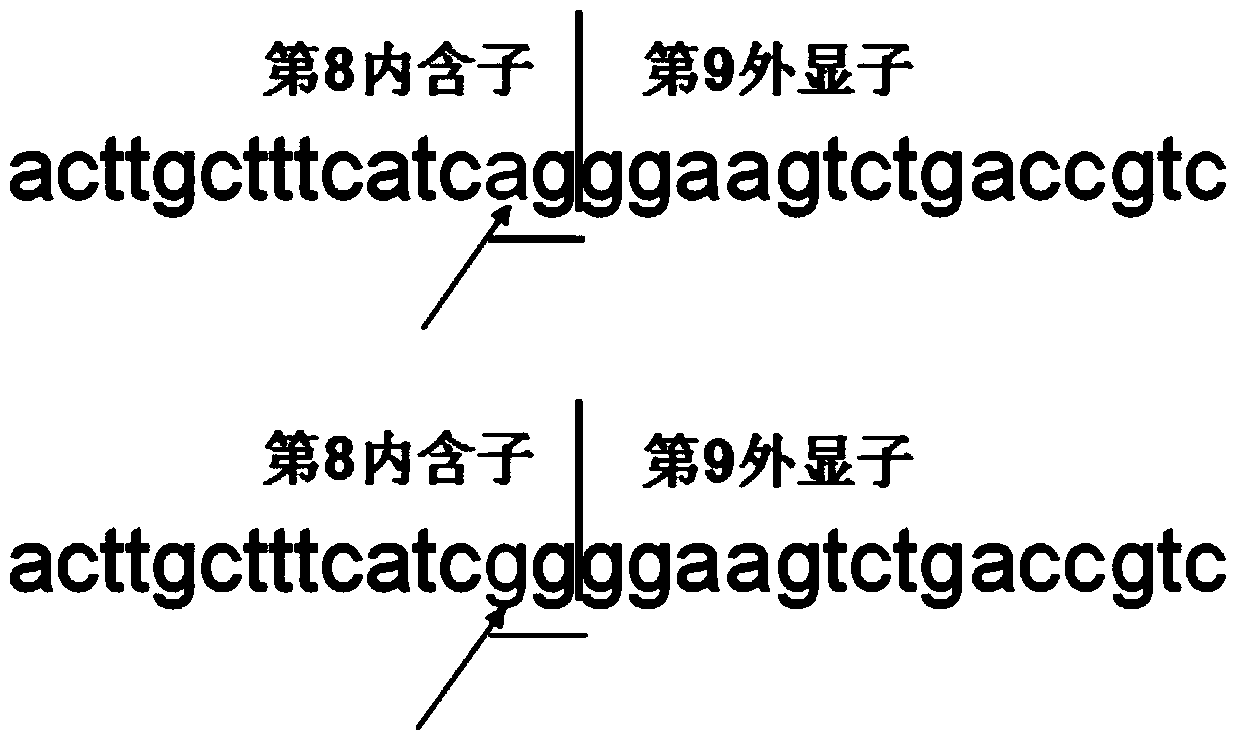
[0092] The PCR products obtained in Example 2, ie, CYP4V2-7, CYP4V2-8 and CYP4V2-9, were digested with restriction enzymes respectively, and the restriction enzymes used were CdiI, ApaI and CdiI respectively. The PCR product CYP4V2-7 gene corresponds to the detected mutation c.802-8_810del17insGC, the CYP4V2-8 gene corresponds to the detected mutation c.992A>C, and the CYP4V2-9 gene corresponds to the detected mutation c.1091-2A>G. The base sequence recognized by the restriction endonuclease CdiI is CATCG, and the base sequence recognized by ApaI is GGGCCC. A change in any base in the recognition sequence will result in the recognition of the restriction endonuclease. The restriction endonuclease digestion reaction system was established as shown in Table 6.
[0093] Table 6 restriction endonuclease digestion reaction system
[0094] ingredients
Volume (ul)
Buffer (10×)
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com