Hepatoma cell aptamer sequence and application thereof
A nucleic acid aptamer and liver cancer cell technology, applied in the field of cell biology, can solve the problem of lack of effective therapeutic drugs for chemotherapy of liver cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
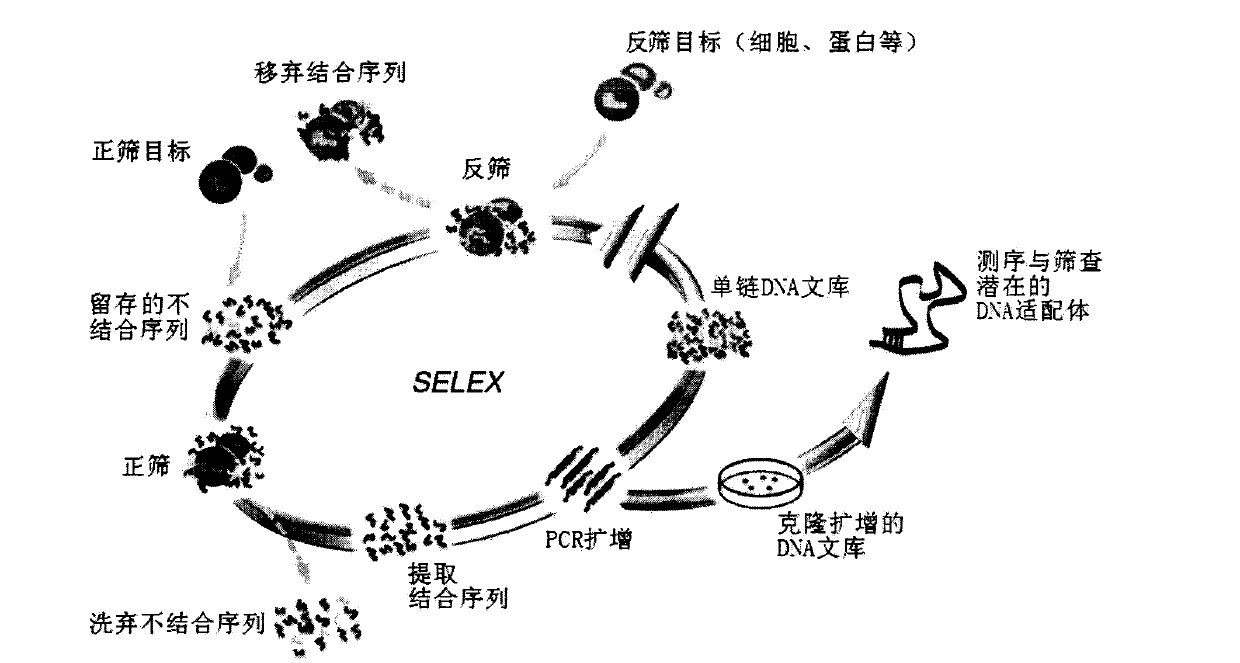
[0017] Example 1 Nucleic acid aptamer screening (one round)
[0018] First round oligo DNA library sequence: 5'-acgctcggatgccactacagNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNctcatggacgtgctggtgac-3'
[0019] Primers used for enrichment:
[0020] Aptamer_L: 5'-FAM-acgctcggatgccactacag-3'
[0021] Aptamer_R: 5'-biotin-gtcaccagcacgtccatgag-3'
[0022] Determination of oligomeric DNA library O D260 Afterwards, it was centrifuged and dried. Dissolve the library with 300ul binding buffer (4.5g / L glucose, 5mMMgCl2, 2mg / mLBSA, 0.2mg / mL yeast tRNA in PBS), take 250pmol (10nmol in the first round), put it on ice at 95°C for 5min, and put it on ice for a while. Then centrifuge quickly.
[0023] Reverse screening: 250pmol of the library was drawn into a 6cm-diameter petri dish with a growth coverage of HL-7702 reaching 70-80%, and made up to 1mL with binding buffer. Shake at 4°C for 1 h. Take the supernatant.
[0024] Positive sieve: Add the supernatant of the reverse sieve into a 6c...
Embodiment 2
[0028] Example 2 Nucleic Aptamer TA Cloning
[0029] Prepare the following solutions in a microcentrifuge tube, the total amount is 5 ul: pMD19-TVector 1 ul, aptamer PCR product 1 ul, dH2O 3 ul. Add 5ul of SolutionI. React at 16°C for 30 minutes. The whole amount (10ul) was added to 100ul DH5a competent cells, and placed in ice for 30 minutes. After heating at 42°C for 45 seconds, place in ice for 1 minute. Add 890ul LB medium, shake and culture at 37°C for 60 minutes. Cultivate on LB agar plate medium containing X-Gal, IPTG, and Amp to form a single colony.
Embodiment 3
[0030] Colony PCR and sequencing of embodiment 3 nucleic acid aptamer clone
[0031] pMD19-T colony PCR and sequencing primers:
[0032] RV-M: 5′-GAGCGGATAACAATTTCACACAGG-3′
[0033]M13(-47): 5'-CGCCAGGGTTTTTCCCAGTCACGAC-3'
[0034] Use a sterilized toothpick to pick white monoclonal colonies and add them to 4ml LB (Amp50ng / L) liquid medium, shake the bacteria overnight at 180rpm at 37°C ( The bacterial liquid was used as a PCR template for PCR amplification. Water was used as a negative control.
[0035] Configure the PCR system: bacteria solution Primer RV-M (10uM) 0.5ul, primer M13(-47) 0.5ul, 2×TaqMix 7.5ul, water 5.5ul (total system 15ul). Amplify on a PCR machine under the following conditions: pre-denaturation at 95°C for 5 min; denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, a total of 33 cycles; total extension at 72°C for 10 min.
[0036] After the reaction, take 3-6ul bacterial liquid PCR product and carry out agarose gel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com