SNP for predicting the sensitivity to anticancer targeted therapeutic formulation
A targeted therapy and sensitivity technology, applied in testing pharmaceutical preparations, testing organic pollutants in water, testing water, etc., can solve problems such as unsatisfactory research results and difficulty in securing a large number of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1: The first stage of screening SNP genes
[0087] 118 patients with colorectal cancer were analyzed by highly aggregated high-speed SNP (using Affymetrix SNP Array 6.0) and in vitro tumor reactivity analysis, and the correlation analysis of these two results obtained 4163 SNPs with a -Log(p) value of 3 or more, from which Screen out 56 cross-inconsistency rates (R 2 ) 0.8 or more SNPs, thereby obtaining the basic information of the sensitive SNP genes of anti-cancer targeted therapy agents.
Embodiment 2
[0088] Example 2: The second stage of screening SNP genes
[0089] Among the SNP genes screened in the first stage above, the candidate SNPs with a frequency of more than 10% in the existing Japanese and Chinese analysis data (www.hanpmap.org) and linkage disequilibrium (Linkage disequilibrium) were screened Genes were additionally screened through non-synonymous, haplotype-tagging and functional SNPs to discover 15 candidate SNPs ( figure 1 ).
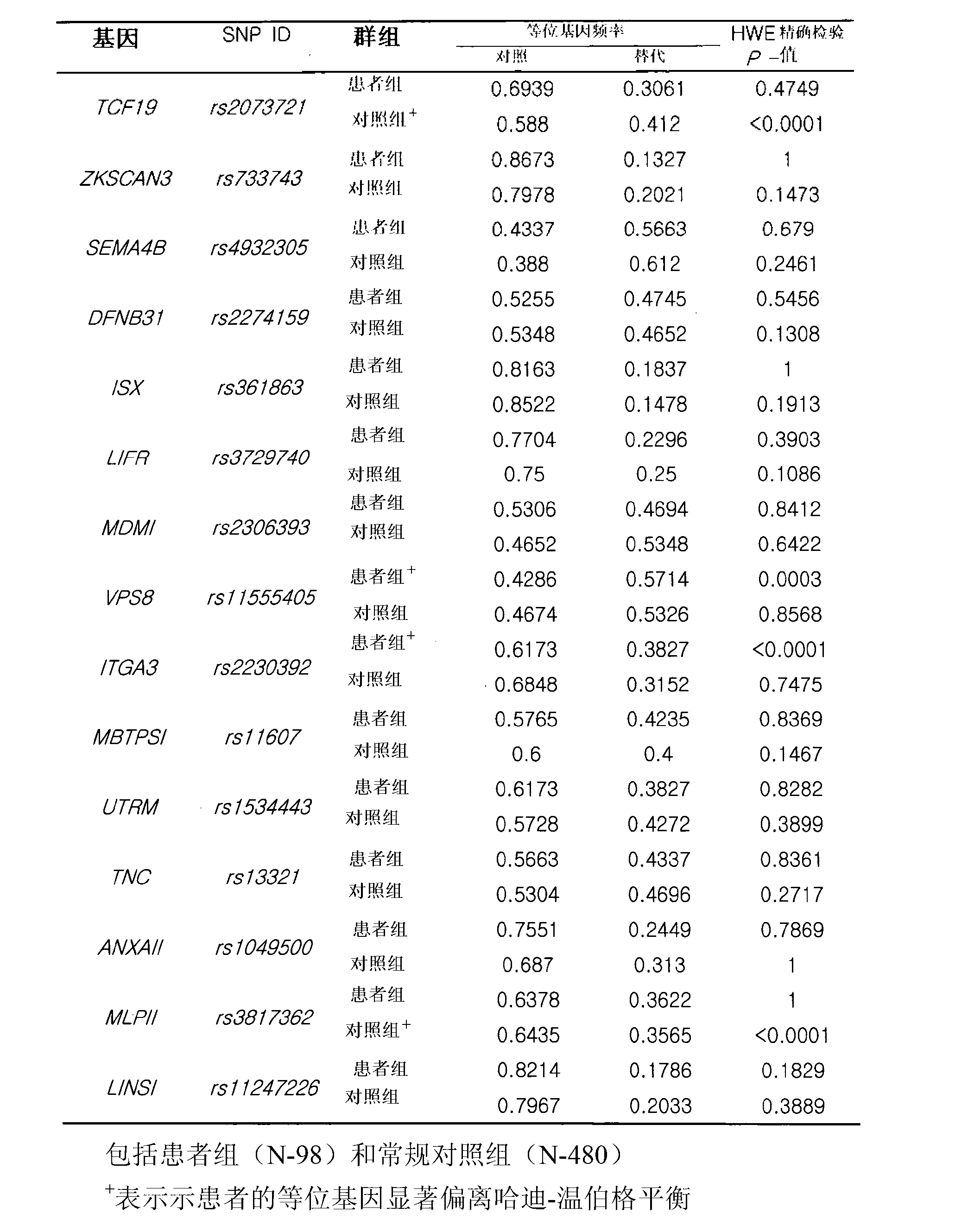
[0090] Then, 480 normal population lymphocyte DNA samples were used to identify the same SNP gene. In the identification results, 11 SNPs that were not significantly (p>0.01) out of the Hardy-Weinberg equilibrium were screened out as Candidate SNPs identified in the clinical association analysis performed for final characterization in Example 3 below (Table 1).
[0091] Table 1. Eleven candidate anticancer agent responsive candidate SNP markers among the eleven genes excavated through two stages of identification
[0092]
[009...
Embodiment 3
[0094] Example 3: Phases of association analysis between SNP genes and existing clinical processes
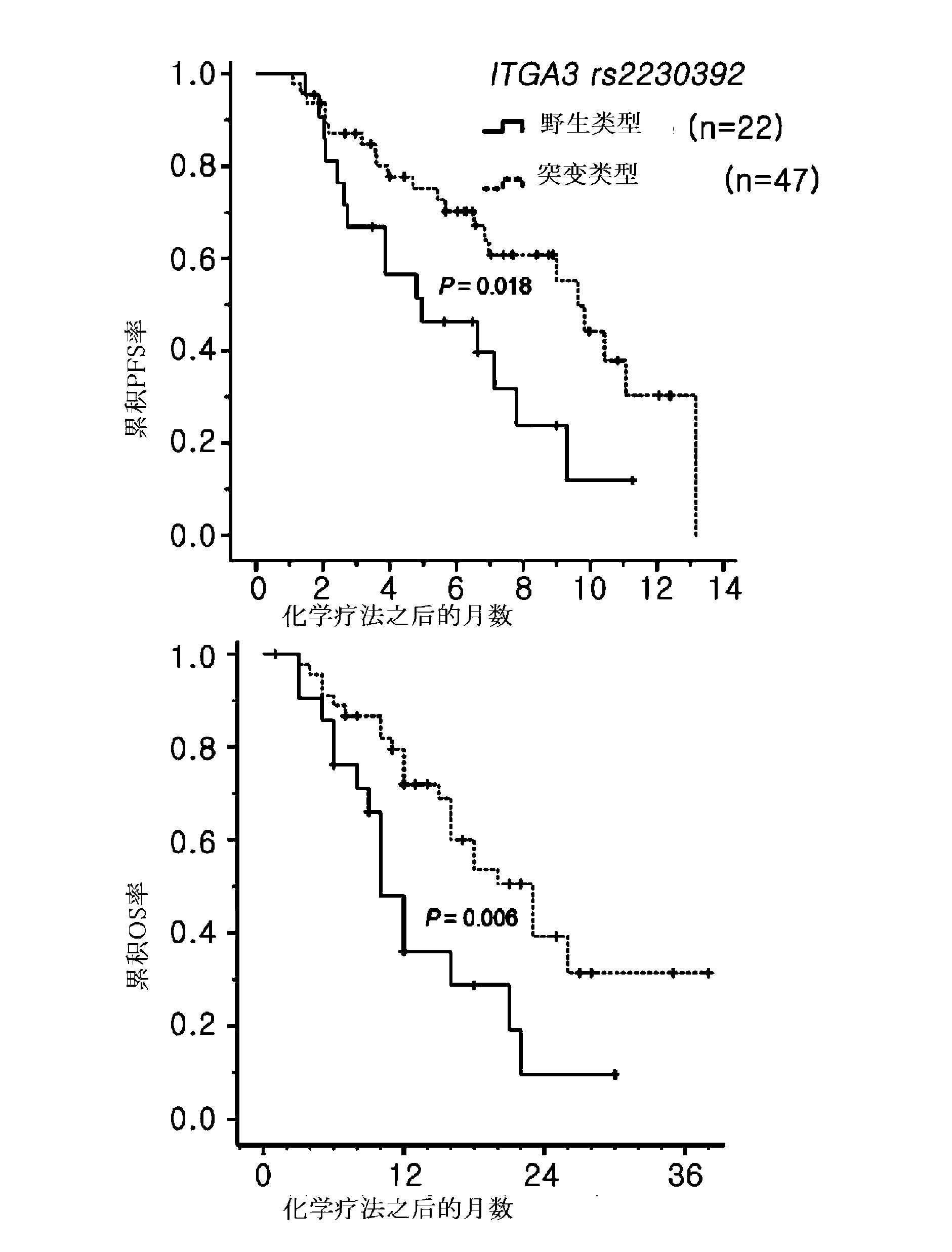
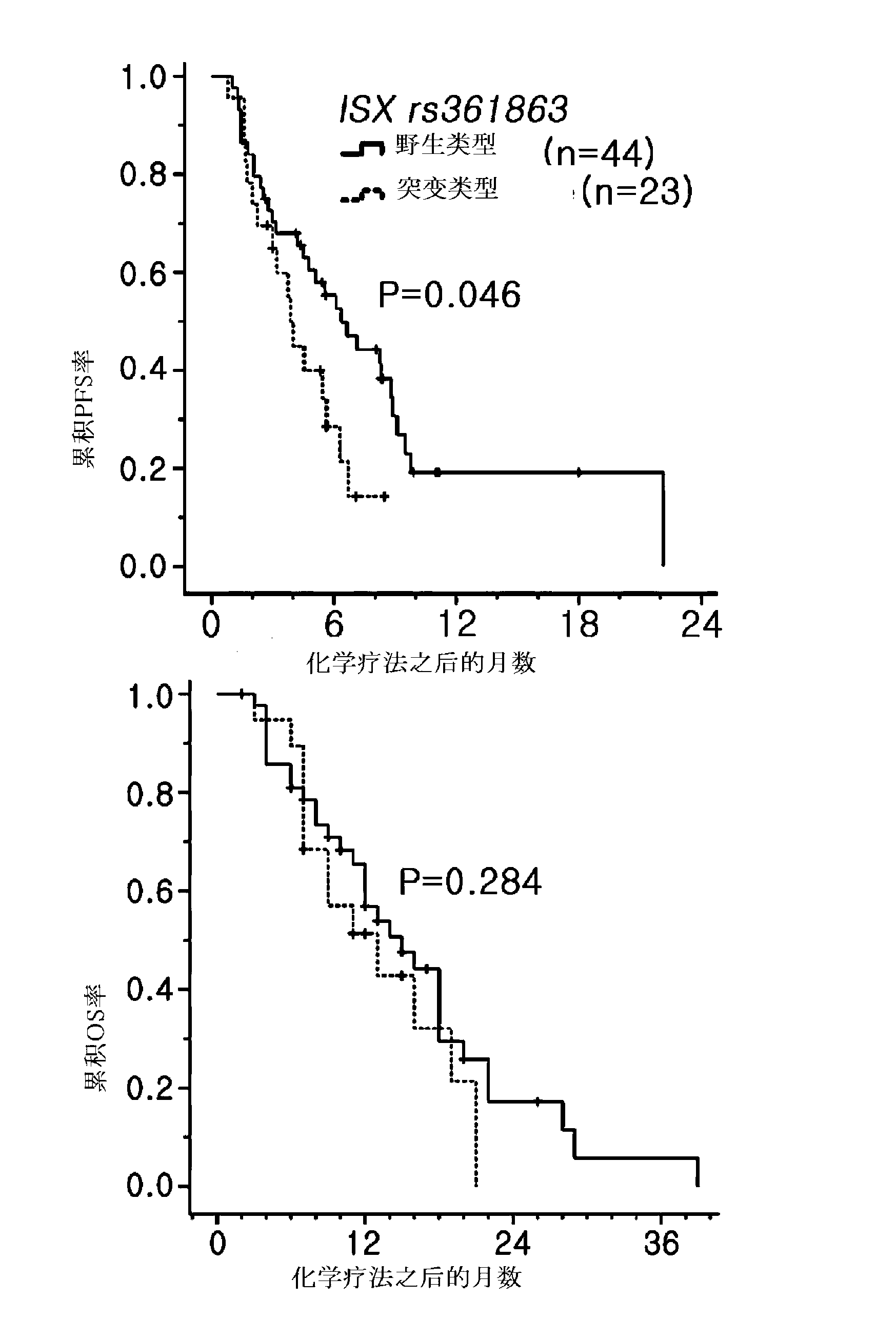
[0095] Clinically, 69 cases of bevacizumab use group and 67 cases of cetuximab use group were used as the research objects, and the clinical correlation analysis was carried out. The patient's clinical treatment process was tracked through the NCCN surveillance guideline (NCCN surveillance guideline) ( www.nccn.org ) was measured, and the tumor reactivity of anticancer agents was determined using RECIST criteria (Therasse et al., J Natl Cancer Inst 2000:92:205-16). The average follow-up observation period of patients is 13 months (range: 1-39 months). Since all the subjects of this preparation are patients with metastatic or recurrent cancer, the follow-up observation period of 13 months should be sufficient for judging the results time.
[0096] 3.1 Survival-related clinical correlations based on the anticancer targeted therapy preparation group analyze
[0097] Corr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com