Method for validating influence of disulfide bond on thermal stability of xylanase
A xylanase and thermal stabilization technology, which is applied in the field of bioengineering, can solve problems such as unseen thermal stability analysis, and achieve the effect of accurate and reliable experimental basis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Homology modeling to simulate the spatial structure of AEx11A:
[0024] Homologous comparison was performed in the PDB database, and the primary structure homology with AEx11A was found to be higher, which were derived from Penicillium funiculosum (1TE1), E.coli (2VUL) and Hypocrea jecorina (1ENX) Crystal structure of family 11 xylanases as a template for homology modeling. Homology modeling of AEx11A was performed using SALIGN (http: / / salilab.org / DBAli / ?page=tools_&action=f_salign) and MODELLER 9.9 (http: / / salilab.org / modeller / ) programs. Through the homology modeling of the AEx11A structure and its alignment with the AoXyn11A structure, it was found that a disulfide bond (Cys 5 -Cys 32 ). The cysteine at position 5 was mutated to threonine (C5T) by site-directed mutagenesis, and the disulfide bond was removed to explore its effect on the thermal stability of AEx11A. The genes before and after the mutation were AEx11A and AEx11A C5T .
Embodiment 2
[0025] Example 2 Mutant gene AEx11A C5T and construction of expression plasmids
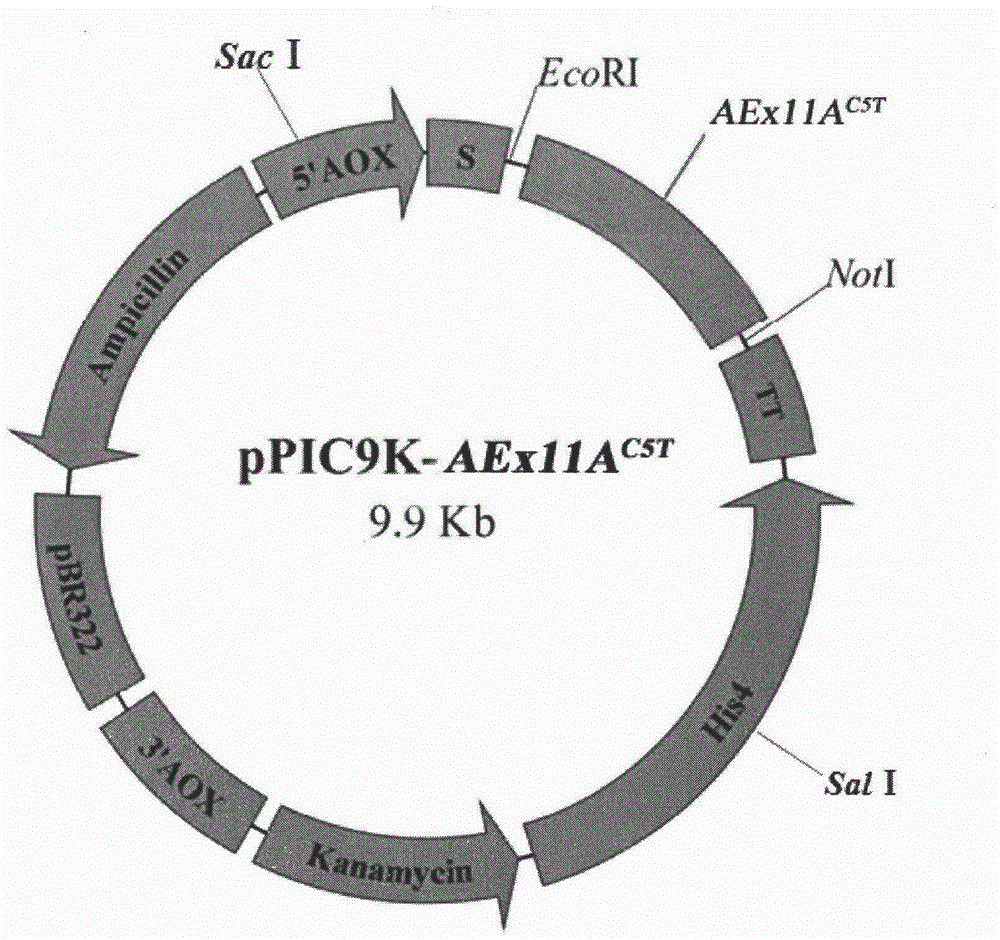
[0026] Design primers based on pPIC9K-AEx11A (this experiment is preserved)
[0027] F1: 5′- GAATTC AACGCTCAAACTactCTTACCTCT-3′, containing EcoR I restriction site and mutation point Thr 5
[0028] R1: 5'- GCGGCCGCT CAATAAACAGTGATAATAGCAG-3′, with Not I restriction site
[0029]The pPIC9K-AEx11A stored in our laboratory was used as a template, and F1 and R1 were used as primers for PCR reaction. 94°C for 2min; 30 cycles, 94°C for 30s, 55°C for 30s, 72°C for 12s; 72°C for 10min. The PCR product was analyzed by 1% agarose gel electrophoresis, and the target band was recovered by tapping the gel and ligated with pUCm-T; transformed into JM109, and sent to Shanghai Sangon for correct sequencing of pUCm-T-AEx11A after enzyme digestion and identification C5T Both the pPIC9K plasmid and the pPIC9K plasmid were digested with EcoR I and Not I, and the recovered digested products were ligated unde...
Embodiment 3
[0030] Embodiment 3GS115 / AEx11A C5T Construction, expression, product purification and activity determination of
[0031] pPIC9K-AEx11A with Sal I C5T Perform linearization, perform electrotransformation and screening according to the Pichia expression manual, and obtain high-copy Pichia recombinant GS115 / AEx11A C5T ; The engineered bacteria were induced with 1.0% methanol for 72 hours, and the mutant xylanase activity in the fermentation broth was basically consistent with that of AEx11A as measured by DNS method. The supernatant after centrifugation of the fermentation broth is the mutant AEx11A C5T The crude enzyme solution was concentrated by an ultrafiltration membrane with a molecular weight cut-off of 10kDa, and then purified by DEAE-Sepharose Fast Flow ion exchange chromatography and Sephadex G-75 gel filtration chromatography. After purification, a single band was detected by SDS-PAGE, And it shows that the molecular weight of the mutant xylanase is basically the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com