Marking method for resistance gene homologues based on EST(Expressed Sequence Tag) data mining
A technology of disease resistance gene and marker method, which is applied in electronic digital data processing, special data processing applications, instruments, etc., can solve the problems of difficult to obtain homologous sequences, difficult to obtain NBS flanking sequences, etc., to speed up the breeding process, type Rich and numerous effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
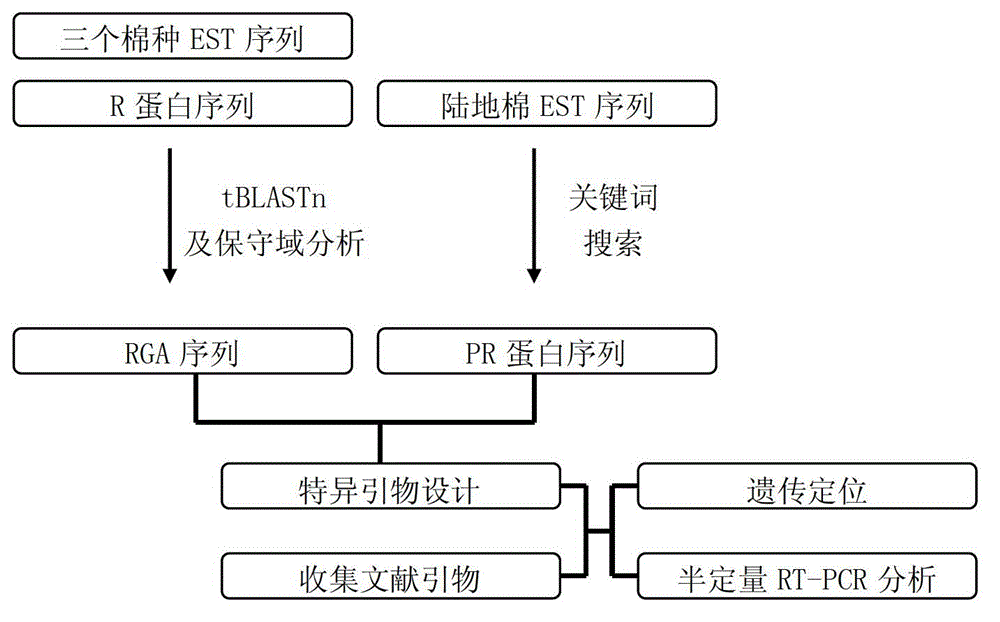
[0029] A method for labeling disease-resistant gene homologues based on EST data mining, the steps of which are:
[0030] (1) Process-related protein (PRP) sequence mining and primer design:
[0031] First download the EST sequence of upland cotton (G.hirsutum) from TIGR (http: / / plantta.jcvi.org / ). Since the sequences in this database have been annotated by genes, several keywords (PR, Pathogenesis and Pathogenesis-related protein) that appear most frequently in PR sequences are directly used to search and obtain PR sequences. After manual exclusion of non-target sequences and redundant sequences, 100 PR sequences were obtained. Since PR sequences often exist in gene families, in order to improve primer specificity, primers were designed using Primer-BLAST (http: / / blast.ncbi.nlm.nih.gov / Blast.cgi), and corresponding specific primers. For the convenience of the test operation, the optimal annealing temperature was uniformly set to 55°C; due to the SSCP technology (Hayashi. P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com