Method of detecting methylated DNA in sample
A technology for detecting samples and methylation, which is used in biochemical equipment and methods, determination/inspection of microorganisms, etc., to achieve the effect of simple detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0037] Preferred embodiments of the present invention will be described below with reference to the drawings.
[0038] The "CpG site" in the present specification refers to a site where cytosine (C) and guanine (G) are adjacent to each other in this order from the 5' to the 3' direction in the base sequence of DNA. In addition, the character "p" of CpG represents a phosphodiester bond between cytosine and guanine.
[0039] The "methylated CpG site" in the present specification refers to a CpG site modified by methylation at the 5-position of cytosine. That is, at the methylated CpG site, 5-methylcytosine (methylated cytosine) and guanine are adjacent in this order from the 5' to the 3' direction.
[0040] "Methylated DNA" in this specification refers to DNA containing at least one 5-methylcytosine.
[0041] In the method of detecting methylated DNA in a sample of the present invention (hereinafter also referred to as "detection method"), first, a sample that may contain meth...
Embodiment 1
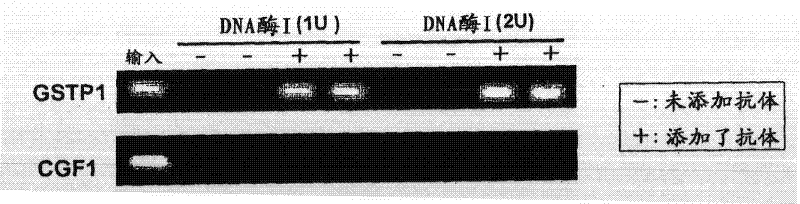
[0085] As the methylated DNA to be detected, 6MeCG oligonucleotide, which is an oligonucleotide containing six 5-methylcytosines, was used. The base sequence of the 6MeCG oligonucleotide is shown below.
[0086]
[0087] 5'-CGAGGTCGACGGTATTGATm5cGAGTATm5cGATAGTm5cGATATm5cGATATm5cGATATm5cGATATACAACGTCGTGACTGG-3' (SEQ ID NO: 1)
[0088] ("m5c" in the base sequence indicates 5-methylcytosine.)
[0089] (1) Sample preparation
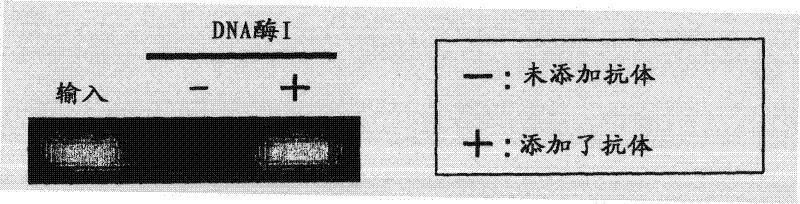
[0090] An aqueous solution of 10 pM 6MeCG oligonucleotide was prepared as a 6MeCG oligonucleotide solution. The 6MeCG oligonucleotide solution was denatured by heating at 95° C. for 10 minutes, and then left to stand on ice for 1 minute. 2 µl was taken from the denatured 6MeCG oligonucleotide solution as an input sample. The remaining solution was used as a sample provided in the detection method of the present invention.
[0091] (2) Contact between sample and anti-methylated DNA antibody
[0092] The above-mentioned sample was divided into two, an...
Embodiment 2
[0113] (1) Sample preparation
[0114] A solution of 6MeCG oligonucleotide was prepared as in Example 1, from which the input sample and the sample provided for the detection method of the present invention were obtained.
[0115] (2) Contact with anti-methylated DNA antibody
[0116] From the above samples, as in Example 1, a test sample and a control sample were obtained.
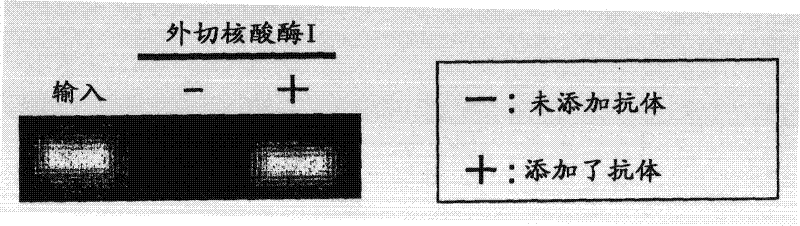
[0117] (3) Decomposition of DNA by deoxyribonuclease
[0118] 4 μl of exonuclease I (20 U / μl: NEB Co., Ltd.), one type of deoxyribonuclease, was added to each of the above test samples and control samples, and reacted at 37° C. for 1 hour. After the reaction, exonuclease I was inactivated by heating each sample at 80°C for 20 minutes.
[0119] (4) Detection of methylated DNA
[0120] In order to check whether the 6MeCG oligonucleotide remained in the sample, quantitative PCR was performed as in Example 1.
[0121] The obtained reaction solution was subjected to electrophoresis using 3% agarose gel, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com