Method and kit for detecting mutation of BRAF gene of human colorectal cancer
A technology for colorectal cancer and human detection, applied in the fields of biotechnology and medicine, can solve problems such as large limitations, and achieve the effects of simple operation, low detection cost, and accurate genotyping
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Selection of samples and extraction of genomic DNA
[0058] The collected genomic DNA comes from whole blood, cells, fresh tissue, and paraffin-fixed tissue. The extraction method refers to the operation manual provided by the kit, and some optimizations have been made at the same time.
[0059] Whole blood genomic DNA of healthy people was used as the wild-type control of the experiment, and was extracted using the BloodGen Mini Kit of Kangwei Century;
[0060] Genomic DNA derived from the colo201 cell line was used as a control for the V600E homozygous mutant type (1799T>A) of exon 15 of the BRAF gene in the experiment,
[0061] The genomic DNA derived from the HT29 cell line was used as the control of the V600E heterozygous mutant type (1799T>A) of exon 15 of the BRAF gene in the experiment, and was extracted using the DNA FlexGenDNA Kit of Kangwei Century;
[0062] Both fresh tissue and paraffin tissue were derived from mice, and were used to evaluate th...
Embodiment 2
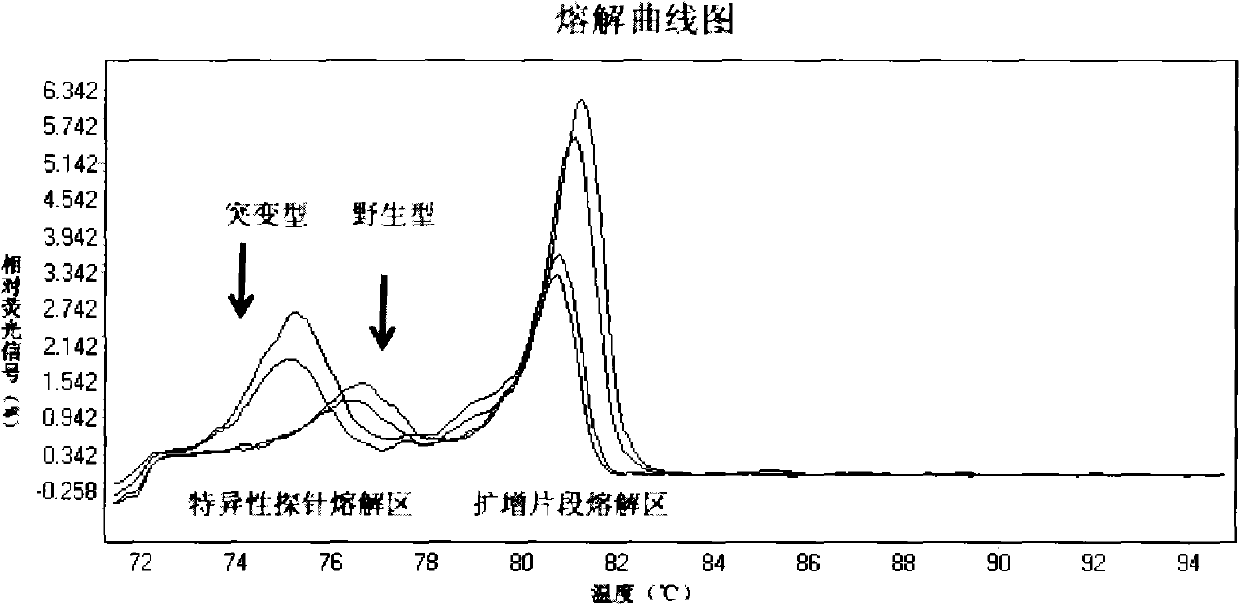
[0068] Example 2: Identification of mutation sites
[0069] HRM technology was used to detect the samples with known BRAF gene exon 15 V600E codon genotype.
[0070] 1. Specific primers and probes are as follows:
[0071] SEQ ID No: 3, Forward primer:
[0072] 5'-ACAACTGTTCAAACTGATGGGACC-3'
[0073] SEQ ID No: 4, Reverse primer:
[0074] 5'-TCCTTTACTTACTACACCTCAGATAT-3'
[0075] SEQ ID No: 2, specific probe: 5'-TCTAGCTACAGTGAAATCTCGAT-3'
[0076]2. Amplify a fragment near the V600E codon by PCR; prepare a mixture: add 1 μl of the genomic DNA solution prepared before, 2 μl of PCR buffer (10×), 1.6 μl of dNTP, 0.1 μl of Taq DNA polymerase, and 0.4 μl of forward primer and 0.08 μl of reverse primer, 2 μl of SYTO 9 fluorescent dye, and PCR-grade pure water to make the reaction volume 20 μl. Reaction at 95°C for 15 minutes, 95°C for 15 seconds, 60°C for 15 seconds, 72°C for 15 seconds, for 20 cycles, 82°C for 15 seconds, 60°C for 15 seconds, 72°C for 15 seconds, for 15 cycles...
Embodiment 3
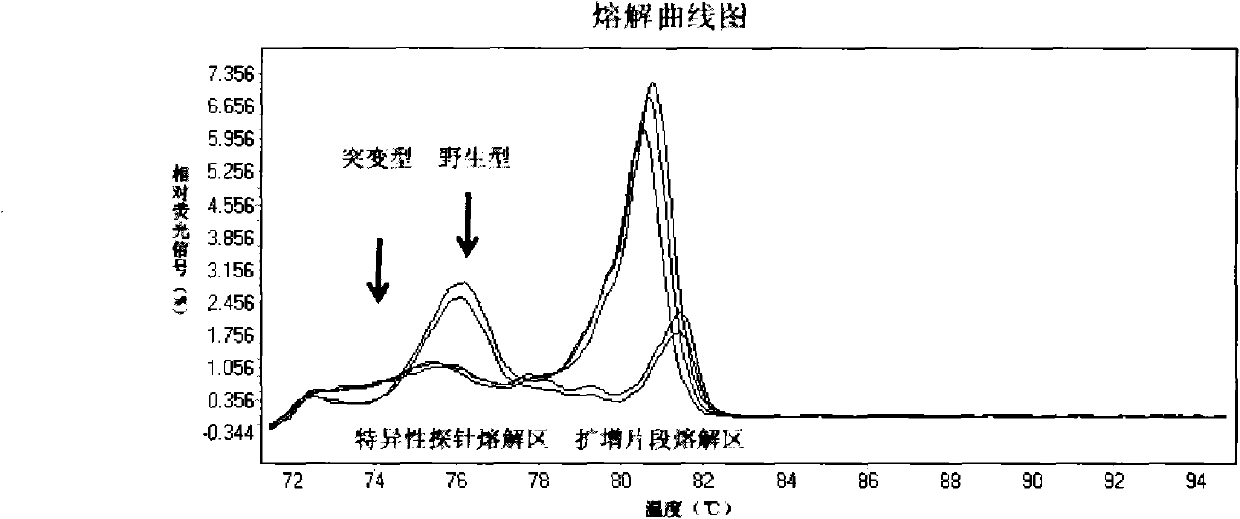
[0080] Example 3: Kit Sensitivity Verification
[0081] The homozygous mutant colo201 genome and the wild-type genome extracted from whole blood were mixed in a certain proportion, so that the homozygous mutant genome accounted for 0.05% of the total DNA amount. The concentration of the above mixed genomic DNA and wild-type genomic DNA was adjusted to 10 ng per microliter.
[0082] 1. Specific primers and probes are as follows:
[0083] SEQ ID No: 3, Forward primer:
[0084] 5'-ACAACTGTTCAAACTGATGGGACC-3'
[0085] SEQ ID No: 4, Reverse primer:
[0086] 5'-TCCTTTACTTACTACACCTCAGATAT-3'
[0087] SEQ ID No: 2, specific probe: 5'-TCTAGCTACAGTGAAATCTCGAT-3'
[0088] 2. Amplify a fragment near the V600E codon by PCR; prepare a mixture: add 1 μl of the genomic DNA solution prepared before, 2 μl of PCR buffer (10×), 1.6 μl of dNTP, 0.1 μl of Taq DNA polymerase, and 0.4 μl of forward primer and 0.08 μl of reverse primer, 2 μl of SYTO 9 fluorescent dye, and PCR-grade pure water to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com