Method for preparing ethanol by fermenting jerusalem artichoke through recombinant saccharomyces cerevisiae
A technology of recombinant Saccharomyces cerevisiae and Jerusalem artichoke, which is applied in the field of ethanol fermentation, recombinant Saccharomyces cerevisiae fermentation ethanol, and the construction of recombinant Saccharomyces cerevisiae, which can solve the problems of poor ethanol tolerance and long fermentation time, and achieve low residual sugar and high sugar alcohol conversion rate , The effect of fast ethanol fermentation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Construction of Inulinase Integrated Expression Vector
[0031] According to the sequence of inulinase (GenBank: X57202), design primers p-inu forward primer: GCCggatccGAATTCTCAAACCGAA (BamH I) and p-inu reverse primer: CGG gcgcgc AGATCAGATCAAACG (BssH II), using K. marxianus Kluyveromyces marxianus (CGMCC2.1549) genome as a template to amplify the p-inu gene. PCR system: 1 μl template, 1 μl each F / R primer, 1 μl enzyme, 4 μl dNTP, 5 μl buffer, ddH 2 O 37μl; PCR program: 94°C pre-denaturation for 5min; 94°C for 1min, 63.6°C for 1min, 72°C for 3min, 30 cycles; 72°C for 10min. After the PCR product was recovered and purified by gel, it was connected with the vector pMD19-T. The positive clones were sequenced after extracting the plasmids, and the results of sequencing analysis proved that the inulinase gene was consistent with the inulinase gene reported by NCBI. pMD19T / p-inu digested with BamH I and BssH II and HO-2 (HO-poly-KanMX 4 -HO, Warren P V, James ...
Embodiment 2
[0032] Example 2 Transformation of S. cerevisiae Saccharomyces cerevisiae (CGMCC2.1882) and verification of transformants .cerevisiae Saccharomyces cerevisiae (CGMCC2.1882) competent cells on YPD (anhydrous glucose 20, yeast extract powder 10, peptone 20 and 1.5% agar powder) plates containing 300 μg / mL G418. Transform by electric shock method, you can refer to BIORAD MicroPulser TMElectric Rotator Instruction Manual. Cultivate at 30°C for 2-3 days, select transformants, and extract the genomic DNA of HI6 / 6. It was verified by PCR method. The primer sequences used for the verification of HI6 / 6 are HI6 / 6 forward primer: AGCTTAATTATCCTGGGCACGAGT and HI6 / 6 reverse primer: GCGAGCTCAGCTCGTTTTCGACACTGGA. PCR program: 94°C pre-denaturation for 5 minutes; 94°C for 1 minute, 60°C for 1 minute, 72°C for 3 minutes, 30 cycles; 72°C for 10 minutes. 1% agarose electrophoresis showed that a fragment of the correct size was obtained, which proved that a positive transformant was obtained....
Embodiment 3
[0033] Example 3 Investigation of Transformant Enzyme Production
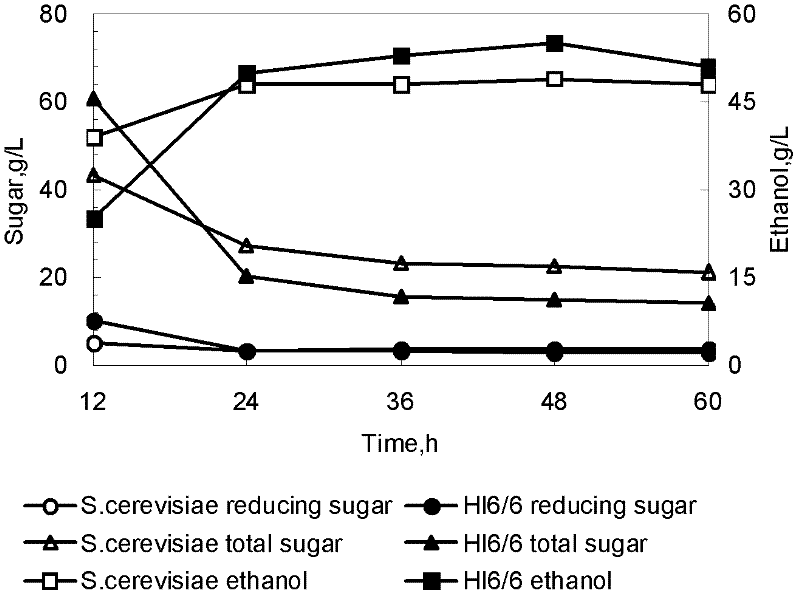
[0034] Take 100 μL of each of S.cerevisiae 6525 and transformant HI6 / 6, inoculate into 50 mL of YPD medium, culture at 30°C for 24 hours, inoculate 10% into seed medium, 100 / 250 mL shake flask, culture at 30°C and take samples every 24 hours . After the fermentation broth was properly diluted, the cell density (OD 600 ); after the fermentation broth was centrifuged at 5000rp for 10 min, the supernatant was used to measure inulinase activity. The biomass of transformant HI6 / 6 was almost the same as that of the starting strain, reaching 9g / L; the inulinase activity of transformant HI6 / 6 reached the highest enzyme activity of 86U / mL at 144h, which was 4.5 times that of the starting strain.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com