DNA (Desoxyribonucleic Acid) ligand for specific binding with EPO (erythropoietin), preparation method thereof and application thereof
An aptamer, specific technology, applied in the field of protein new recognition element-DNA aptamer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1. Construction of random single-stranded DNA (ssDNA) library and its primers
[0078] (a) Construct a random ssDNA library with a length of 80 bases:
[0079] 5'-CTTCTGCCCGCCTCCTTCC-N 39 -GGAGACGAGATAGGCGGACACT-3' wherein N represents any one of the bases A, G, C, T (see Berezovski M, MusheevM, Drabovich A, Krylov SN.Non-SELEX Selection of Aptamers.J.Am.Chem.Soc.2006 , 128:1410-1411), the capacity of the library is 10 13 -10 15 .
[0080] (b) Synthesize the forward primer labeled with fluorescein (FAM) at the 5' end:
[0081] 5'-FAM-CTTCTGCCCGCCTCCTTCC-3' (SEQ ID NO: 121),
[0082] (c) Synthesize the reverse primer of the 5' terminal labeling biotin (biotin):
[0083] 5'-Biotin-AGTGTCCGCCTATCTCGTCTCC-3' (SEQ ID NO: 122).
[0084] The random ssDNA library and primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd.
Embodiment 2
[0085] Example 2. Screening of DNA aptamers that specifically recognize rHuEPO-α
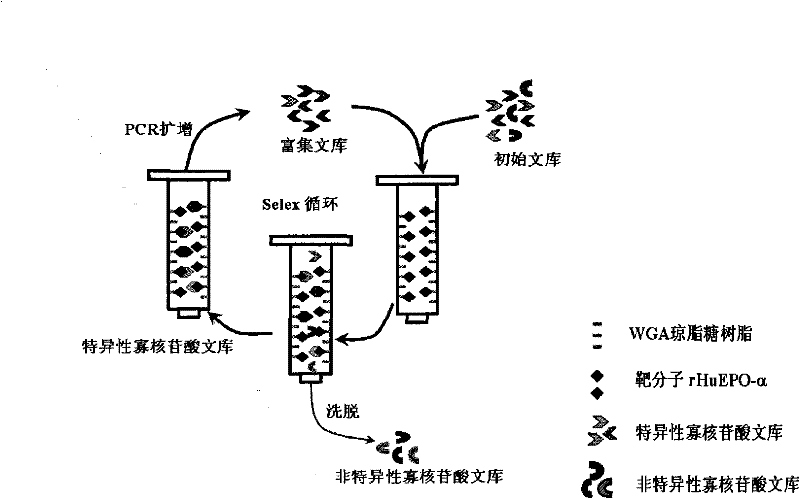
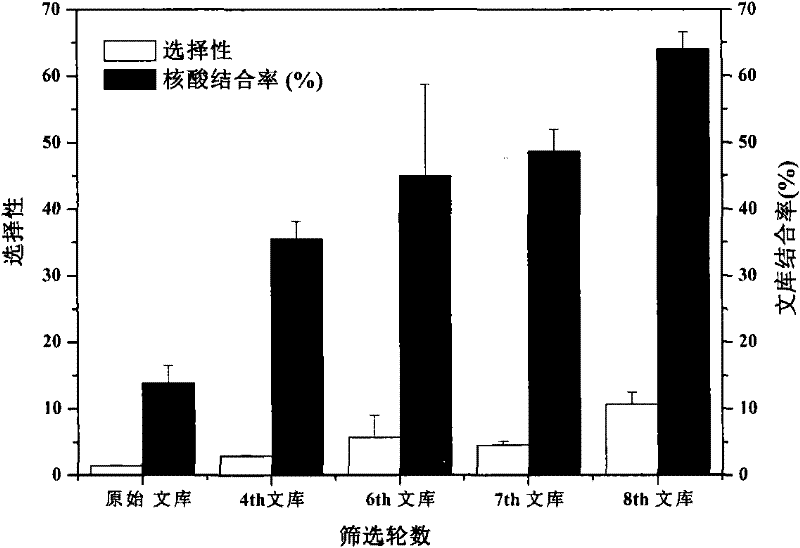
[0086] In order to screen out DNA aptamers with strong binding specificity to rHuEPO-α, a total of 8 rounds of screening were carried out by the SELEX method (8 SELEX cycles, see figure 1 ), the following takes the first round of screening as an example to describe this process in detail.
[0087] (1) SELEX method to screen the ssDNA library that specifically binds to rHuEPO-α
[0088] (a) Reverse screening treatment of wheat germ agglutinin coupling resin and ssDNA library
[0089] Take 2000 pmol of the random ssDNA library and dissolve it in 300 μL 1×SELEX screening buffer (1×SELEX screening buffer: 20 mM Tris-HCl, 140 mM NaCl, 5 mM MgCl 2 , 5mM KCl, PH=7.4), and placed in a water bath at 95°C for 5 minutes, then immediately placed in an ice bath for 10 minutes.
[0090] Then 220 μL of wheat germ agglutinin (WGA)-coupled agarose resin (particle size 200-300 μm, purchased from Sigma-Aldr...
Embodiment 3
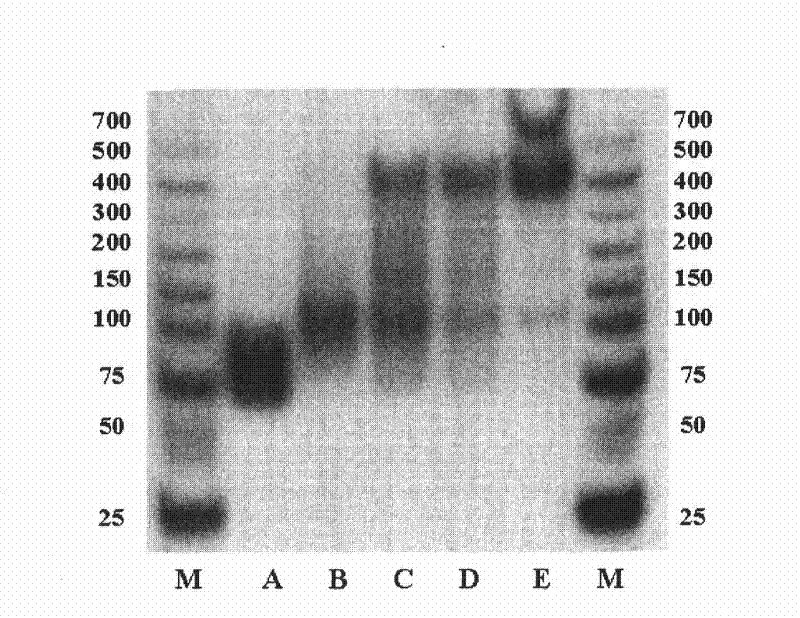
[0143] Example 3. Electrophoretic mobility shift assay (EMSA) of screened oligonucleotide libraries
[0144] (1) Radioisotope labeling of ssDNA library
[0145] Add 2 μL of labeling buffer (10×T4 polynucleotide kinase buffer, TaKaRa Company), 30 pmol of ssDNA to be labeled with radioisotope (original ssDNA library and oligonuclei obtained from the 4th, 6th, and 8th rounds of screening) to the EP tube. nucleotide library), 4 μL water, 20 U (activity unit) of T4 polynucleotide kinase, 20 μL Ci[γ- 32 P]ATP, make up the volume to 20 μL with water, mix well, and incubate at 37°C for 1 hour. Immediately act at 68°C for 10 minutes to inactivate the activity of the enzyme, and filter the solution with a 30kDa ultrafiltration tube to remove free [γ- 32 P]ATP, finally dissolved in water to obtain 60μL [γ- 32 P] ATP-labeled ssDNA solution.
[0146] (2) EMSA determination
[0147] Take 25nM of [γ- 32 P] The ATP-labeled ssDNA library was incubated with 0, 25, 50, 75, 100, 200, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com