Method for detecting N-acylated homoserine lactone
A technology for acylated homoserine lactone and sample detection, which is applied in the direction of material analysis by observing the influence on chemical indicators, and analysis by making materials undergo chemical reactions, etc., to achieve high sensitivity, broad application prospects, and good specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1, the preparation of ahyr receptor protein
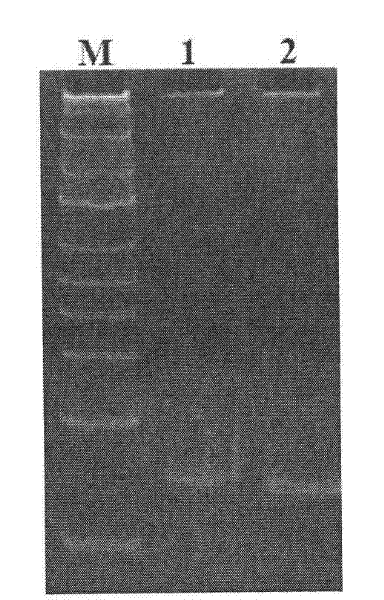
[0053] 1. Extraction of total DNA from Aeromonas hydrophila ATCC 7966 and cloning of the gene of ATCC 7966 receptor protein AhyR
[0054] The total DNA of Aeromonas hydrophila ATCC 7966 was extracted according to the instructions of the bacterial DNA extraction kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China). The primers AhyR-EF (5′-ccggaattca tgaccgttaa aaaactttac-3′) (SEQ ID: 3) and AhyR-ER (5′-attgcggccg cttataaaaa ttcaggaaat g- 3′) (SEQ ID: 4), respectively introduce endonuclease EcoR I and Not I restriction sites (underlined part) at the end of the primer, carry out PCR amplification with total DNA of Aeromonas hydrophila ATCC 7966 as template . The PCR reaction parameters were as follows: denaturation at 94°C for 5 minutes and cooling to 4°C; denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 1 min, and incubation at 72°C for 10 min after 30 cycles. The obt...
Embodiment 2
[0059] Embodiment 2, the application of acceptor protein
[0060] 1. Binding function with target DNA
[0061] 1. The gene recognition DNA (inter) sequence of Ah ATCC 7966 receptor protein AhyR was obtained
[0062] The primers AhyR-BE (5'-CTCGTTTTTCTACCTCCCATC-3') (SEQ ID: 5) and AhyR-BR (5'-CAGTTGGTCTTGTTTCATATGC-3') (SEQ ID : 6), using Aeromonas hydrophila ATCC 7966 total DNA as a template for PCR amplification. The PCR reaction parameters were as follows: denaturation at 94°C for 5 minutes and cooling to 4°C; denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, extension at 72°C for 1 min, and incubation at 72°C for 10 min after 30 cycles. Purified and preserved. The nucleotide sequence of the obtained DNA fragment is shown in SEQ ID NO:7.
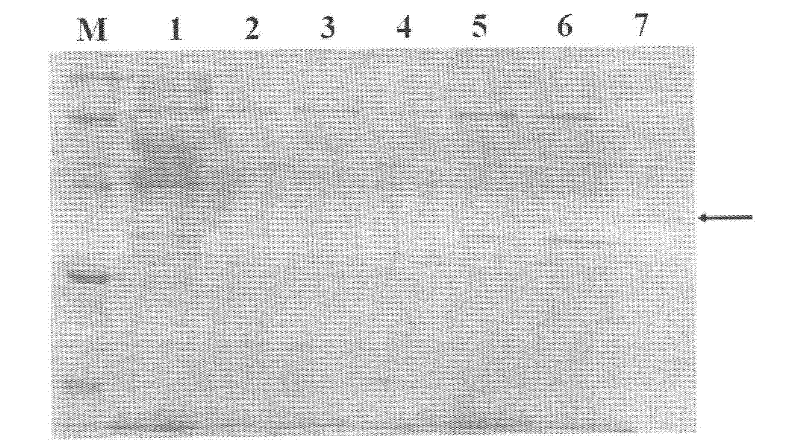
[0063] 2. Gel retardation experiment of the interaction between the signal molecule receptor protein (AhyR) of AhATCC 7966 and its recognition DNA (inter) sequence
[0064] The receptor protein (AhyR) and its recognition...
Embodiment 3
[0075] Example 3, Ah ATCC 7966 signal molecule receptor protein (AhyR) nano-magnetic bead surface modification
[0076] Nanomagnetic beads were purchased from Shanghai Aorun Micro-Nano New Material Technology Co., Ltd. (Shanghai, China), AllMag TM PM3-008.
[0077] 1. Preparation of modified nano-magnetic beads:
[0078] 1. Magnetic beads activated by carbodiimide N-hydroxysuccinimide method (EDC / NHS method)
[0079] (1) Take 2mg PM3-008 magnetic beads into a 1.5ml centrifuge tube, wash twice with 500μl MEST buffer, and remove the supernatant after magnetic separation. The MEST buffer was prepared as follows: 100 ml of MES buffer (2-(N-morpholine)ethanesulfonic acid) with a concentration of 10 mM and pH 6.0 was mixed with 0.05 ml of Tween 20 to obtain a MEST buffer.
[0080] (2) Preparation of EDC solution: Dissolve the EDC solid (1ml: 5mg) in MES buffer with a concentration of 10mM and a pH of 6.0 stored at 4°C to obtain an EDC solution (ready for immediate use);
[0081]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com