Method for identifying Maotai-flavor Daqu liquor
An identification method, Daqu's technology, applied in the direction of microorganism-based methods, microorganism measurement/inspection, biochemical equipment and methods, etc., can solve problems such as differences, differences in the number and population distribution of microorganisms, and highly heterogeneous chemical and physical properties , to achieve accurate results, avoid screening and enrichment, easy separation and identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] 1. Fungal DNA
[0052] 1. Prepare samples
[0053] The Daqu produced in Maotai-flavored Liquor Daqu Production Workshop of Kweichow Maotai Town Jinshi Liquor Co., Ltd. was sampled at different times in a cycle during the koji making process. Sampling in the Daqu production workshop of Jinshi Liquor Industry Co., Ltd.
[0054] 2. Extraction and purification of total DNA
[0055] Take 3-6g Daqu + 25mL phosphate buffer (pH8.0) + 0.1% PVPP, + 1mL soil temperature (80 or 60) shaker for 30min, ultrasonic for 6-7min, let stand for 2-5min, and settle at low speed (1000rpm) Centrifuge for 5min, take out the supernatant, mix, centrifuge at high speed (9000rpm) for 8min, remove the supernatant, then add phosphate buffer (PVPP), 25mL, break up and centrifuge at high speed (9000rpm) for 8min, remove the supernatant.
[0056] Add 10-15mL extract solution (the composition of the extract solution is: 100mM Tris-HCl (tris(hydroxymethyl)aminomethane hydrochloride) pH8.0, 100mM sodium ...
Embodiment 2
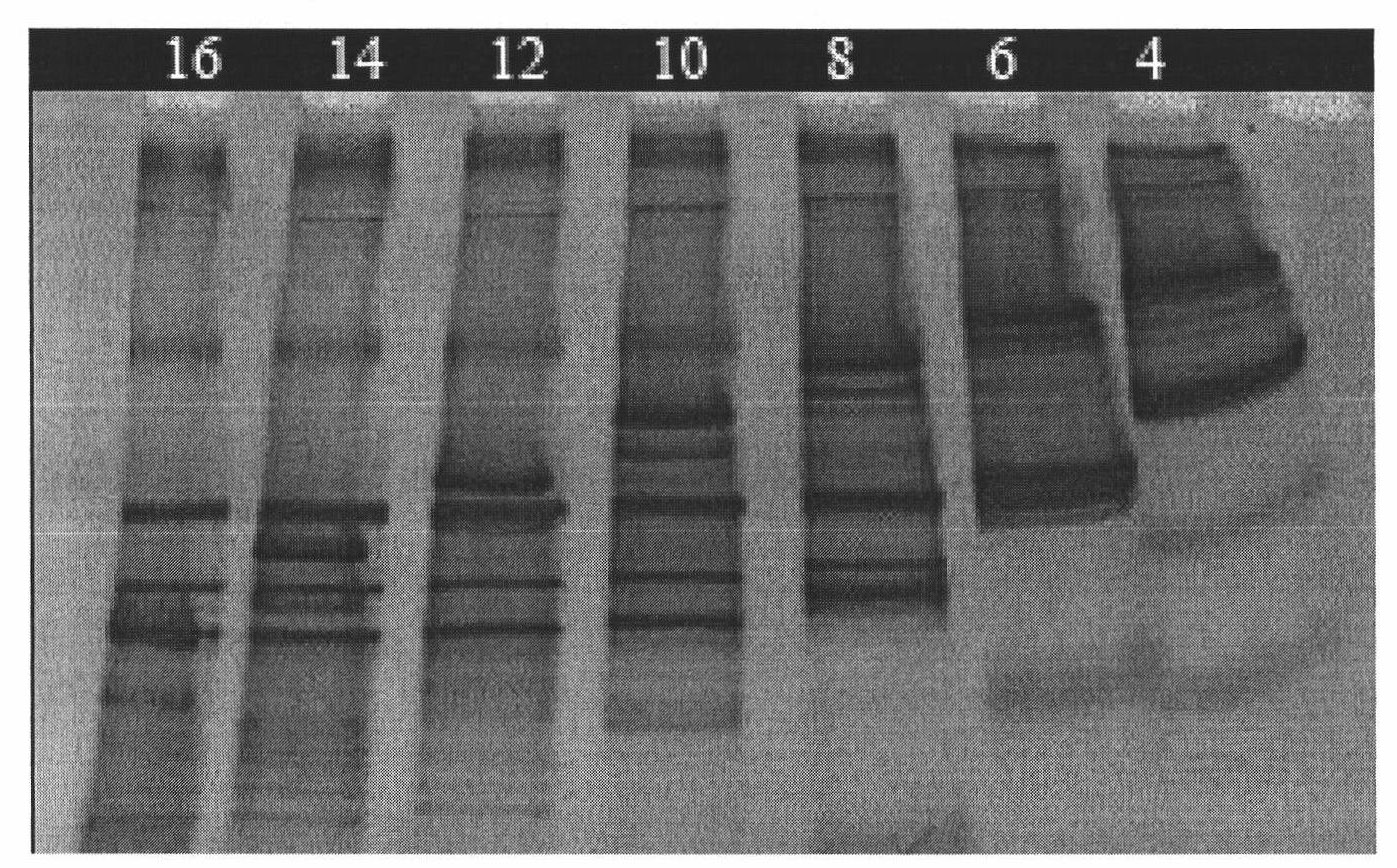
[0120] Embodiment 2, to the analysis of the dominant flora of finished koji bacteria and fungi in the sauce-flavored high-temperature Daqu, the method used is the same as in Example 1, and the spectrum obtained is as follows Figure 5 , the sequence comparison analysis is shown in Table 3
[0121] Table 3 Sequence alignment analysis of the DGGE fingerprint of the finished song analysis corresponding to a single band
[0122] DGGE
band
Accession number
Closest identity
%identity
1
AJ535638.1
Bacillus
galactosilyticus
99
2
AB269425.1
Uncultured bacteria
100
3
AY422988.1
Virgibacillus sp.
99
4
AB211027.1
100
5
DQ225173.1
Thermoactinomyces
sp.
100
6
EU162045.1
100
[0123] It can be seen that the dominant bacterial groups in the finished product Daqu of Maotai-flav...
Embodiment 3
[0125] Embodiment 3, get unknown Daqu sample and measure as blind sample, and used method is identical with embodiment 1, and the spectrogram that obtains is as follows Figure 6
[0126] It can be seen from the spectrum that the spectra of samples H and T are similar, and the finished Maotai-flavored liquor Daqu is consistent with the actual situation. The dominant bacteria are: Uncultured bacteria, Virgibacillus sp., Bacillus sp. and Thermoactinomyces sp.
[0127] The dominant flora of sample L fungi are: Saccharomyces sp., Saccharomycopsis fibuligera strain and Mucor sp. are consistent with the actual.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com