Method for detecting single nucleotide polymorphism of cattle chemerin gene
A single nucleotide polymorphism and detection method technology, which is applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of no yellow cattle, etc., and achieve low-cost and high-precision effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
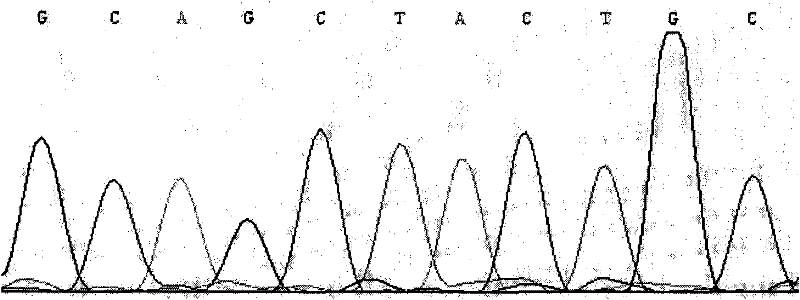
[0036] 1. Design of PCR primers for the region containing the ninth exon of cattle chemerin gene
[0037] Taking the bovine (NC_007302.4) sequence published by NCBI as a reference, Primer 5.0 was used to design PCR primers capable of amplifying the ninth exon region of the ox chemerin gene. The primer sequences are as follows:
[0038] Forward primer: 5'-CGGAGCCCACCAGACAGG-3'18nt;
[0039] Reverse primer: 5'-CAGCGGGCAGCAGCACAC-3'18nt;
[0040] Using the above primers to amplify the cattle genome, it is possible to amplify a 441bp gene fragment including the first exon region of the cattle chemerin gene (NC_007302.4 sequence) and part of the intron 750-1190bp; sequence the amplified fragment After identification, it was found that when the G mutation at the 868th bp (that is, the 12th position of the CDS region of the chemerin gene) was changed to A, the amino acid codon encoding the 4th nt was mutated from CTG to CTA, thereby forming a synonymous mutation of leucine; and Thi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com