Nucleic acid aptamer for classification of different-subtype non-small cell lung cancers and screening method thereof
A nucleic acid aptamer and cell technology, applied in biochemical equipment and methods, individual particle analysis, material excitation analysis, etc., can solve the problems of insufficient specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
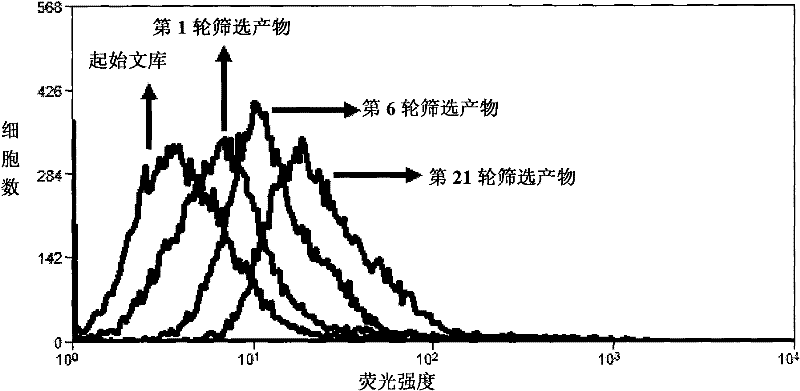
[0044] Example 1. Screening of nucleic acid aptamers for typing different subtypes of non-small cell lung cancer
[0045] 1. Design and synthesis of random nucleic acid library
[0046] Design and synthesize a random nucleic acid sequence library comprising 20 nucleotides at both ends and 45 nucleotides in the middle as follows: 5'-ACGCTCGGATGCCACTACAG(N 45 ) CTCATGGACGTGCTGGTGAC-3'.
[0047] 2. Screening of nucleic acid aptamers
[0048] Dissolve the random nucleic acid library in binding buffer (PBS, 150mM NaCl, 5mM MgCl 2 , 1mg / ml yeast transfer RNA), incubated with lung adenocarcinoma cell line A549 cells on ice for 1 hour; washed with buffer solution (PBS, 150mMNaCl, 5mM MgCl 2 ) after washing, the A549 cells were scraped off, and then the DNA bound to the cell surface was dissociated by heating; the dissociated DNA was incubated with the large cell lung cancer cell line HLAMP cells on ice for 1 hour, and the supernatant was collected and used DNA was used as a templa...
Embodiment 2
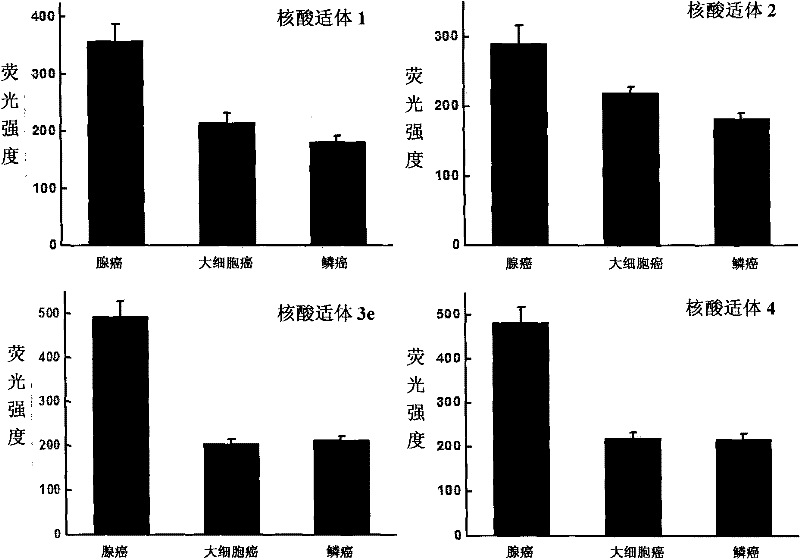
[0061] Example 2. Applying nucleic acid aptamers to type different subtypes of small cell lung cancer
[0062] The four aptamers obtained in Example 1 were labeled with fluorescein isothiocyanate (FITC). The random nucleic acid library of Example 1 was labeled with fluorescein isothiocyanate (FITC).
[0063] Different cultured tumor cells were washed twice with PBS, then treated with 0.02% EDTA for 3 minutes, and washed twice with PBS.
[0064] Each tumor cell was subjected to the following four groups of treatments, and each treatment was repeated three times, and the results were averaged:
[0065] Treatment 1: by cell counting, about 300,000 cells were respectively dispersed in the binding buffer solution, and then fluorescein isothiocyanate (FITC) was added to label the nucleic acid aptamer 1 (the final concentration was 100 nM).
[0066] Treatment 2: by cell counting, about 300,000 cells were collected and dispersed in the binding buffer solution, and then fluorescein i...
Embodiment 3
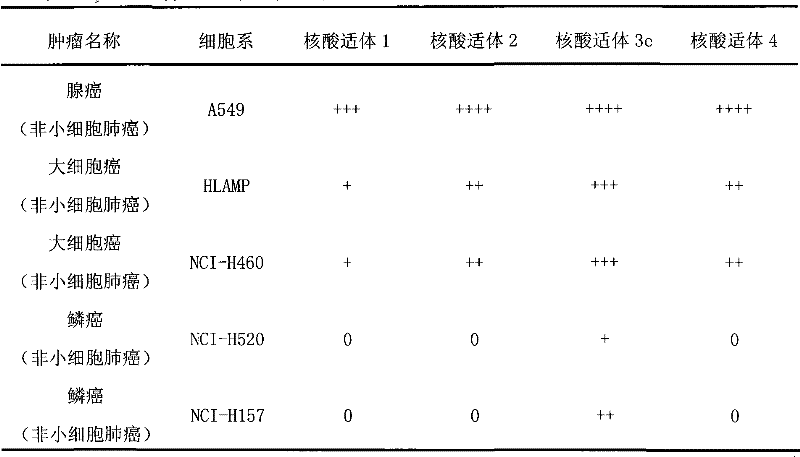
[0075] Example 3. Detection of the binding of the nucleic acid aptamers listed in Table 1 to lung cancer tissue sections
[0076] Formaldehyde-fixed paraffin-embedded tissue sections from different volunteers with lung cancer were dewaxed twice in xylene at room temperature for 20 minutes each time, and then immersed in a series of ethanol for hydration (100%, twice, 1 minute each time) ; 95% ethanol, 1 time, 1 minute; 70% ethanol, 1 time, 1 minute). The hydrated tissue sections were then immersed in buffer solution and heated at 95°C for 15 minutes. After the tissue sections returned to room temperature, they were incubated with binding buffer containing 20% fetal bovine serum and calf thymus DNA for 1 hour at room temperature. After washing, the tissue sections were respectively combined with tetramethylrhodin-labeled nucleic acid aptamers (200nM) Incubate in buffer for 1 hour at room temperature. After staining, the tissue sections were washed 3 times with washing buffe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com