MicroRNA used for inducing leukemia cell differentiation
A molecular, tumor cell technology, applied in the field of double-stranded RNA molecules and their applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Screening of 12 positive siRNAs
[0049] K562 cells were cultured in IMDM medium (purchased from Thermo-Fisher, catalog number SH30228.01B) supplemented with 10% fetal bovine serum (purchased from Sigma, catalog number SH30228.01B) in a cell culture incubator (purchased from Sanyo) at 37°C with 5% carbon dioxide (Unless otherwise indicated, the K562 cell culture method is the same in the following examples), will contain 3x10 7 Random siRNA library mixed plasmids with different siRNA coding sequences were transfected into 6.7x10 7 For K562 cells, 48 hours after transfection, G418 (purchased from Merck, catalog number 345810; final concentration 800ug / ml) was added to the culture medium to kill untransfected cells; after 14 days, the cell concentration was adjusted to 1×10 7 cells / ml, according to every 10 6 Ratio of adding 1ug of antibody to each cell Use anti-CD235 antibody (purchased from R&D, product number MAB1228) to label the cells at 4°C for half a...
Embodiment 2
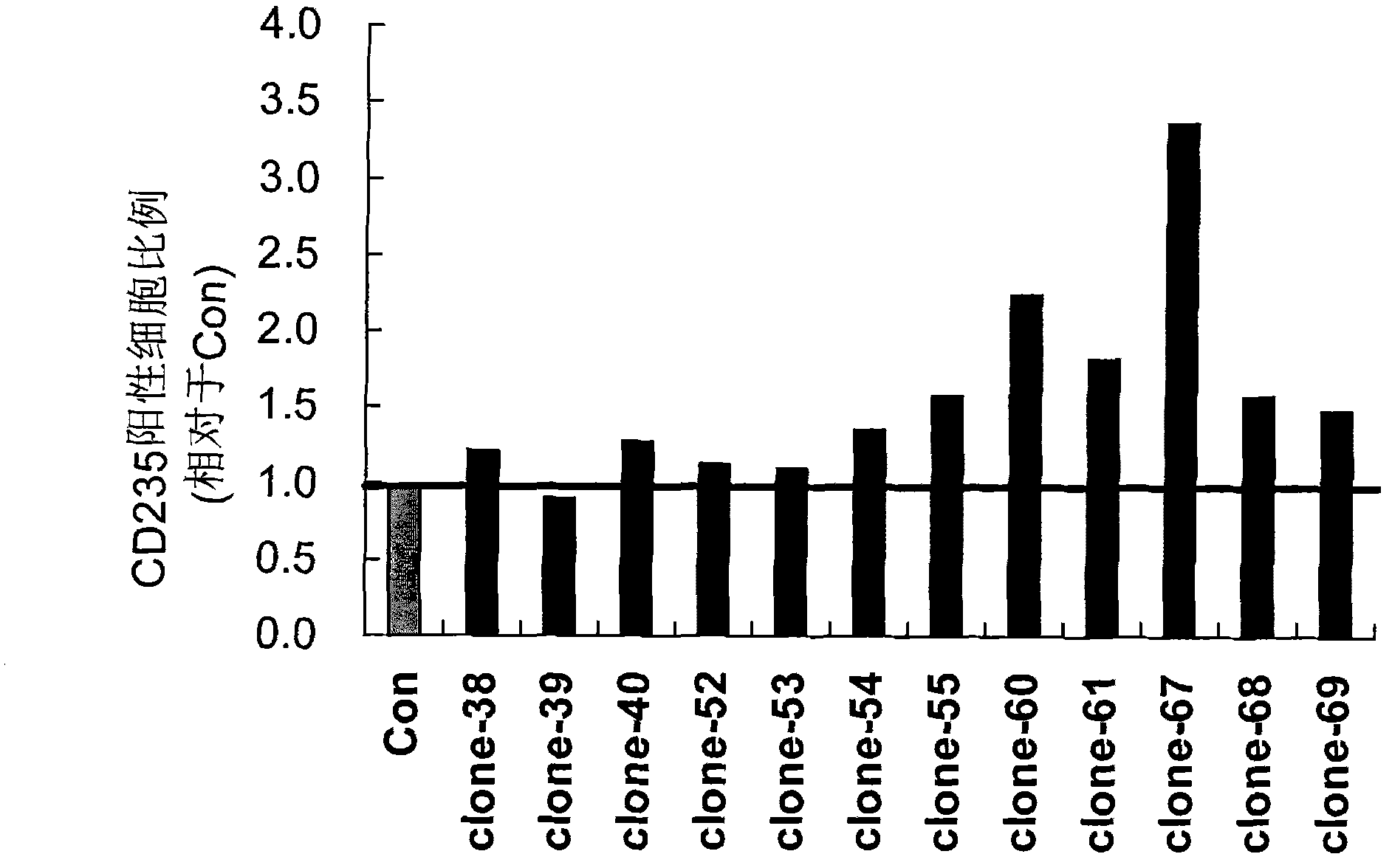
[0051] Example 2: 12 siRNAs increase the expression of CD235 in K562 cells to varying degrees
[0052] In addition to 12 randomly synthesized siRNAs, a negative control group (Con) was set up: only the transfection reagent HiPerFect Transfection Reagent was added during transfection, and no siRNA group was added.
[0053] K562 cells were cultured according to the method of Example 1, and when the cells reached (at a confluence of about 60%), these siRNA duplexes were respectively transiently transfected into K562 cells separately with HiPerFect Transfection Reagent (purchased from Qiagen, Cat#301704), The concentration of siRNA was 25nM during each transfection, and the second transfection was carried out 60 hours after the first transfection, and the cells were collected 48 hours and 72 hours after the second transfection, and were used to detect the indicators of erythroid differentiation, thereby The role of these siRNAs in inducing erythroid differentiation was identified....
Embodiment 3
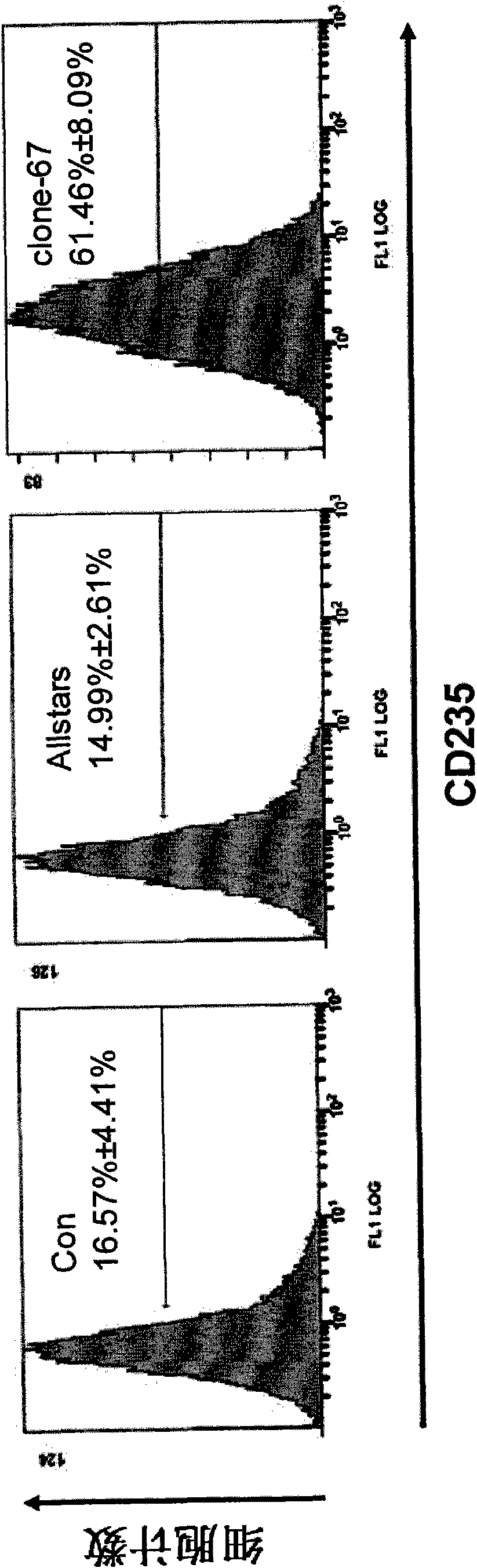
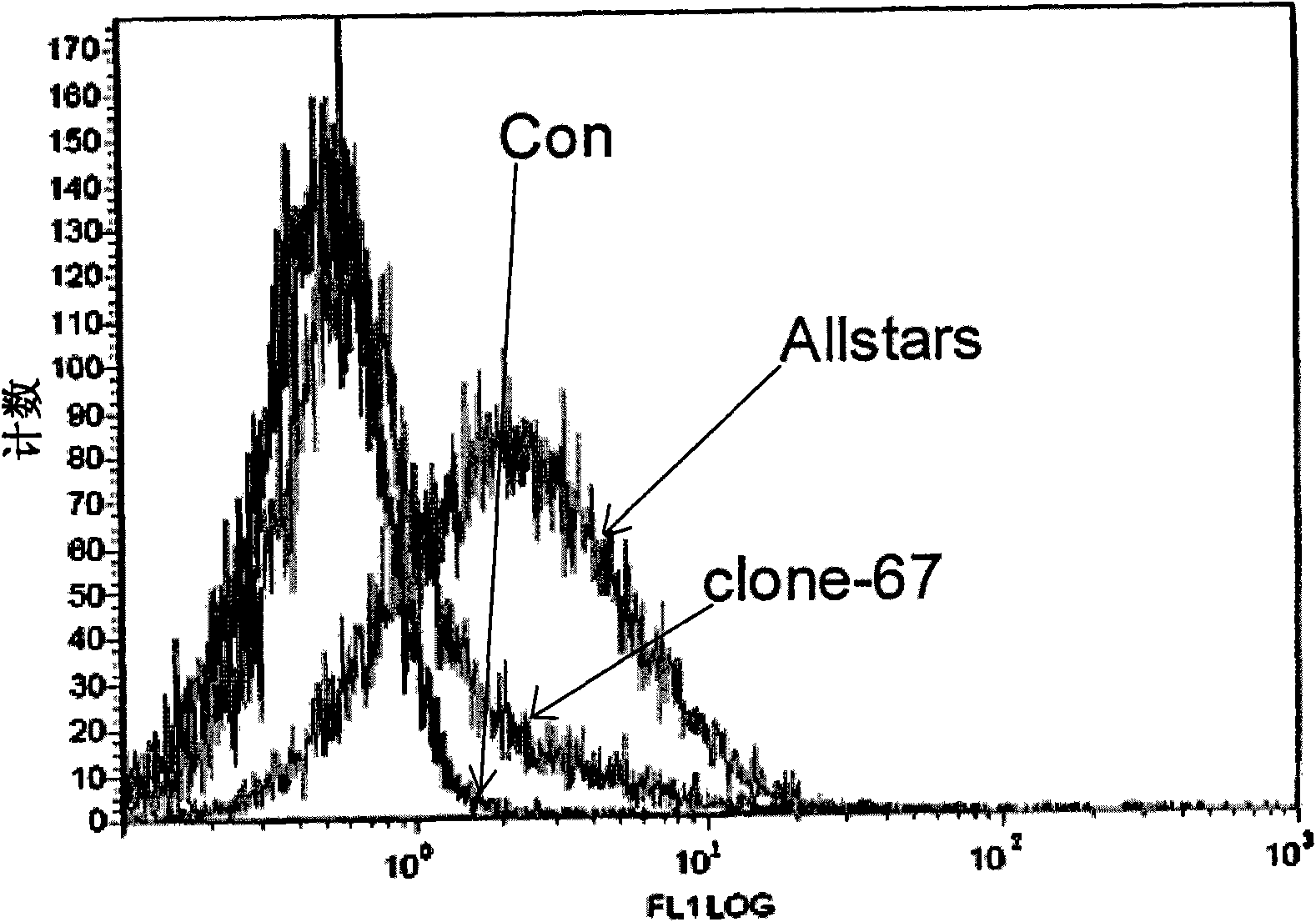
[0056] Example 3: clone-67siRNA increases CD235 expression in K562 cells
[0057] The culture method and transfection method of CD235 cells were the same as in Example 2, except for the clone-67siRNA obtained in Example 2, all identification experiments were set up with two negative control groups (unless otherwise specified, the negative control groups in the following examples were Same as this): transfection only added transfection reagent HiPerFect Transfection Reagent, no siRNA group (Con group); transfection does not act on any gene negative control Allstars negative control siRNA (purchased from Qiagen, Cat#1027280) group (Allstars group).
[0058] Detection of CD235 expression in K562 cells transiently transfected with clone-67siRNA, Con and Allstars: Cells in each group were collected 72 hours after two transfections, and FITC-labeled anti-CD235 antibody (purchased from BD Pharmingen, catalog number 559943) was used at 4°C After staining for 1 hour, the cells were wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com