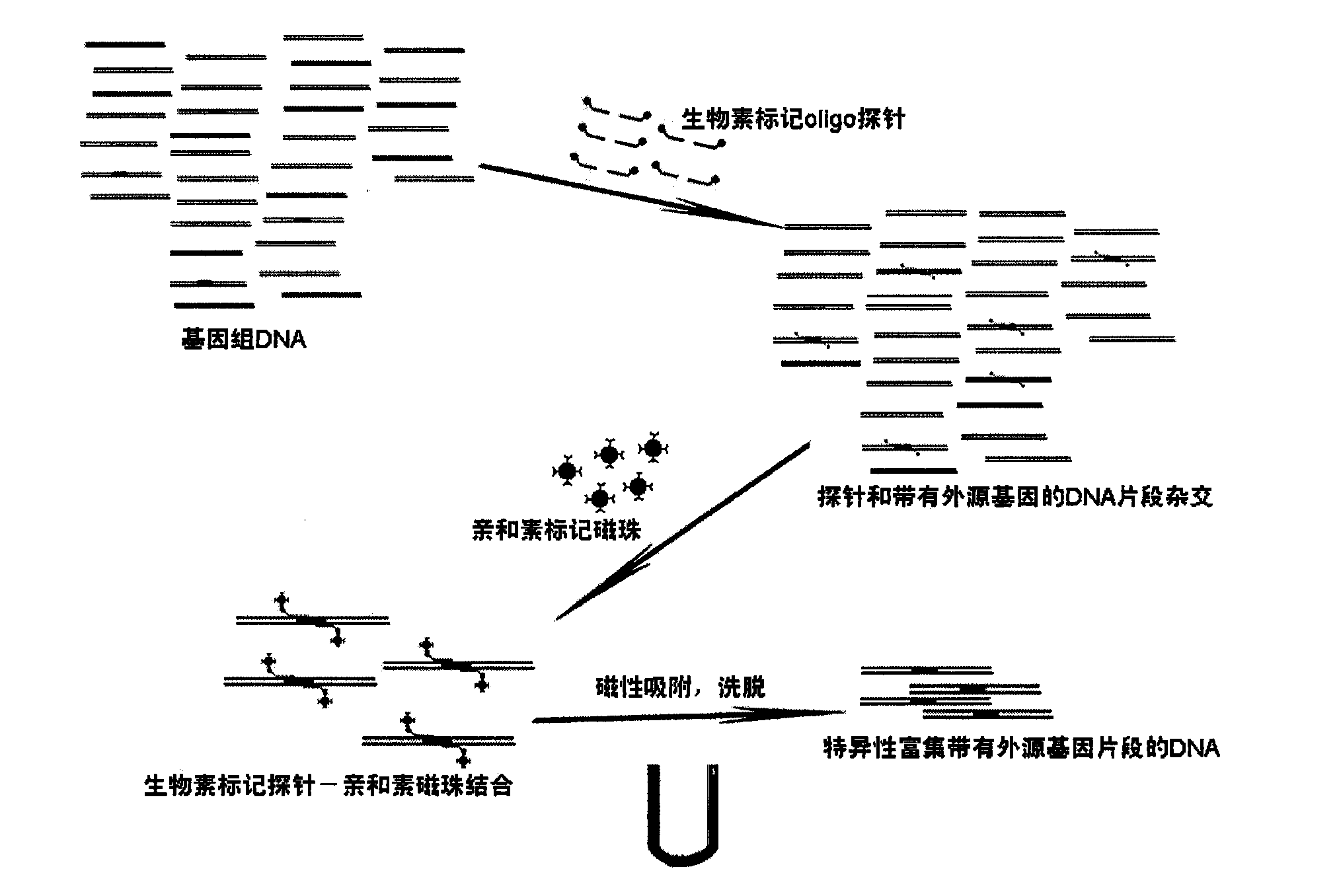
Method for gathering foreign DNA in transgenosis product
A transgenic and product technology, applied in the field of detecting whether a sample contains transgenic components, can solve the problems of low copy number of exogenous DNA and false negatives, and achieve the effects of avoiding false negative results, fast detection time and cheap price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Taking the transgenic soybean (strain: ARG04) produced by Monsanto as the detection object, take the detection of the exogenous gene CaMV35S promoter in the transgenic soybean as an example:
[0048] Primers used in this example:
[0049] 35S-A (upstream primer): 5'-TGGAAAAGGAAGGTGGCT-3' (SEQ ID NO: 1)
[0050] 35S-B (downstream primer): 5'-CTTGCGAAGGATAGTGGG-3' (SEQ ID NO: 2)
[0051] In this embodiment, the biotin-labeled target gene probe detects between the upstream primer and the downstream primer, and its sequence is:
[0052] 5'-CCTACAAATGCCATCATTGCG-3' (SEQ ID NO: 3)
[0053] The exogenous target gene detected is the CaMV 35S promoter, and the promoter sequence is (SEQ ID NO: 4):
[0054] tggaaaagga aggtggctcc tacaaatgcc atcattgcga taaaggaaag gccatcgttgaagatgcctc tgccgacagt ggtcccaaag atggaccccc acccacgagg agcatcgtggaaaaagaaga cgttccaacc acgtcttcaa agcaagtgga ttgatgtgat atctccactgacgtaaggga tgacgcacaa tcccactatc cttcgcaaga cccttcctct atataaggaagttcatttca tttg...
experiment example
[0060] 1. PCR detection of DNA extraction solution:
[0061] Using the sterile aqueous solution of DNA obtained in Example 1 as a template, electrophoresis analysis was performed after PCR amplification. figure 2 To represent the electrophoresis analysis chart of DNA extraction solution PCR detection result:
[0062] from figure 2 As can be seen from the PCR detection results, there is no other amplified gene interference in the PCR reaction (4,5 negative control has no specific band), and the primers used are specific; the genes of non-transgenic soybeans will not be amplified (2,5) 3 has no specific band); the primers used can amplify the target fragment of transgenic soybean (1 has a specific band).
[0063] 2. Detection of the limit concentration (the concentration that the PCR reaction just cannot amplify):
[0064] After multiple dilutions of the sterile aqueous solution of DNA obtained in Example 1, electrophoresis analysis was performed after PCR amplification. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com