Improved production and in vivo assembly of soluble recombinant icosahedral virus-like particles
A recombinant virus, soluble technology, applied in the direction of virus/phage, virus, virus peptide, etc., can solve the problem of icosahedral virus CP-peptide fusion particle assembly and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
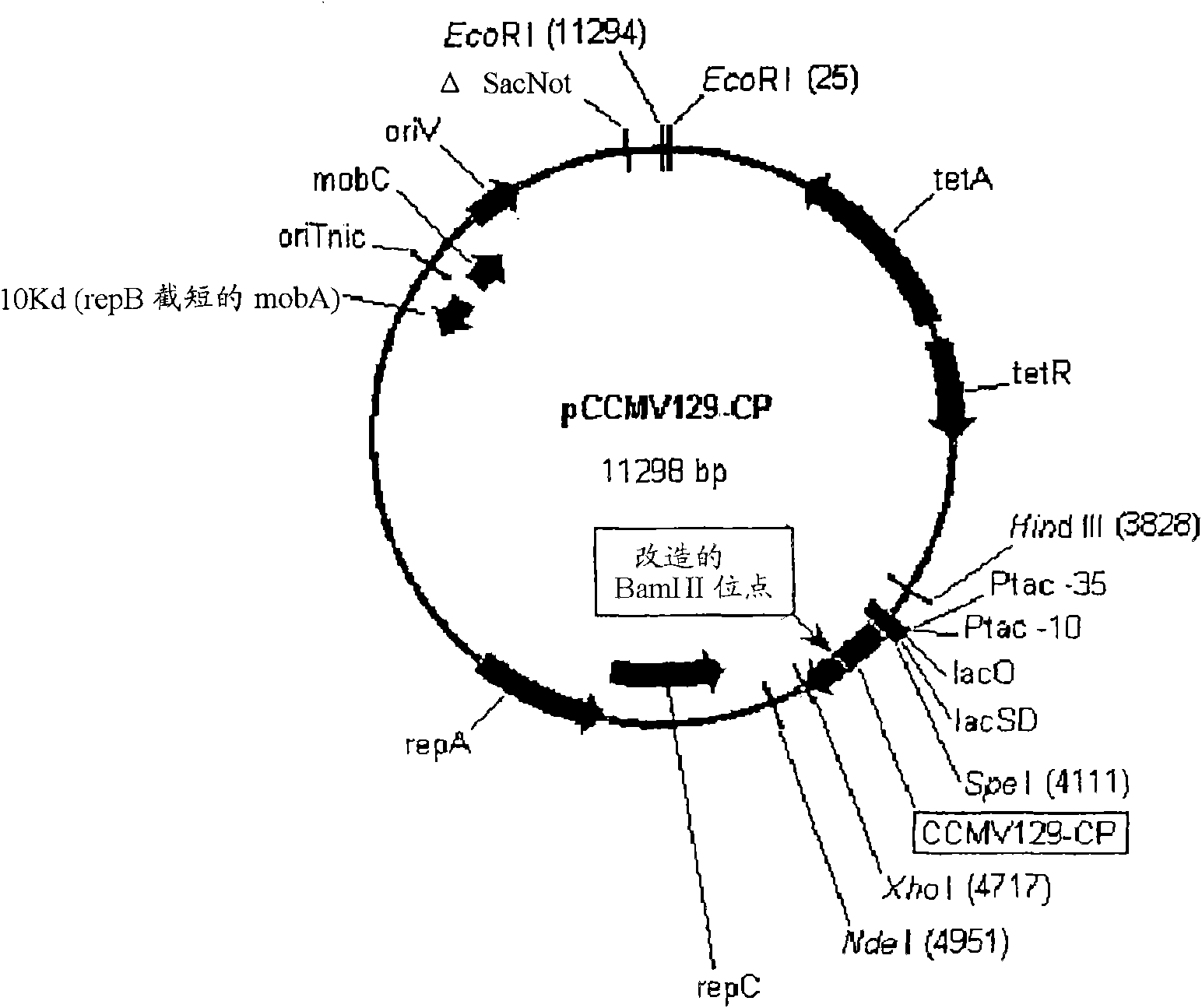
[0318] Example 1: Cloning of expression plasmids for expression of codon- and hydrophilicity-optimized CCMV capsid proteins in Pseudomonas fluorescens
[0319] clone:
[0320]A codon and hydrophobicity optimized CCMV CP nucleotide sequence (SEQ ID NO: 3) was designed. A shuttle plasmid (DNA 2.0, MenloPark, CA). The insert was gel purified on a 1% agarose gel and ligated into vector pDow1169 (a medium copy plasmid with RSF1010 origin, pyyF, tac promoter, and rrnBT1T2 terminator from pKK223-3 (PL-Pharmacia)) to create an expression plasmid (SEQ ID NO: 23) for expression of CCMV CP in Pseudomonas fluorescens. After purification with a Micro Bio-spin 6 column, the ligated product was transformed into Pseudomonas fluorescens strain DC454 (ΔpyrFRXF01414(lsc)::lacIq1) by electroporation. After shaking in LB medium at 30°C for 2 hours, the transformants were plated on M9 glucose plates. The presence of the insert was confirmed by restriction digest and sequencing of plasmid DNA i...
Embodiment 2
[0511] Example 2: Introduction of restriction sites into loops of codon and hydrophilicity optimized CCMV capsid proteins
[0512] Site-directed mutagenesis reactions were performed using Quikchange II-XL (Stratagene, TX) according to the manufacturer's protocol. The Pseudomonas fluorescens expression plasmid (SEQ ID NO: 23) containing the codon-optimized CCMV-CP served as template. After purification with Micro Bio-spin 6, the resulting plasmid with the introduced restriction sites was transformed into Pseudomonas fluorescens strain DC454 (ΔpyrF RXF01414(lsc)::lacIq1) by electroporation. Protein expression was performed as described in Example 1.
[0513] Primers used to introduce the blunt-end cleavage restriction site AfeII into loop 63:
[0514] CCMV-AfeI-63-F (SEQ ID NO: 24): 5'-TGCGCGGCTGCCGAGAGCGCTGCCAAGGTCACCAGT-3'
[0515] CCMV-AfeI-63-R (SEQ ID NO: 25): 5'-ACTGGTGACCTTGGCAGCGCTCTCGGCAGCCGCGCA-3'
[0516] Primers used to introduce the 3' overhang cleavage restrict...
Embodiment 3
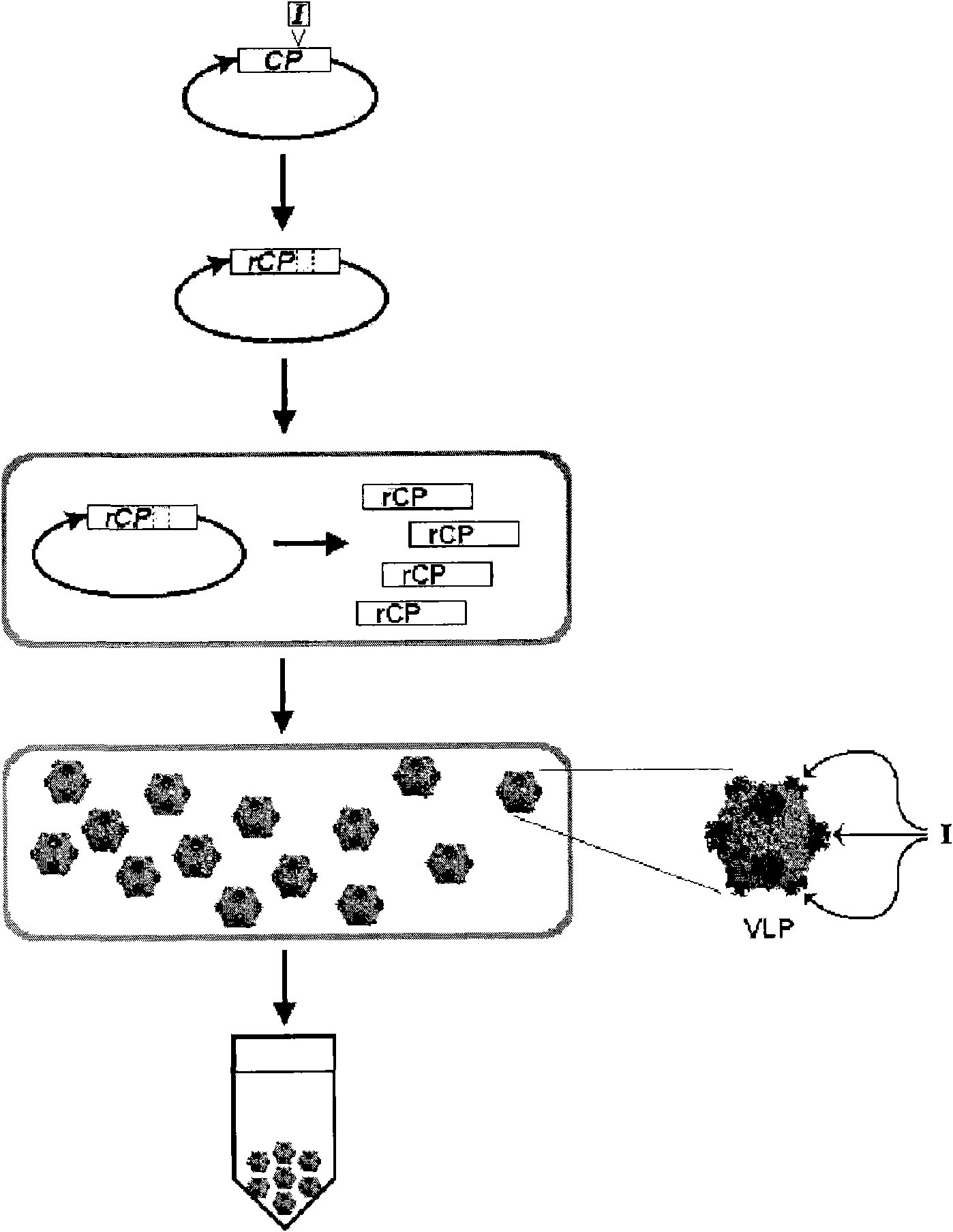
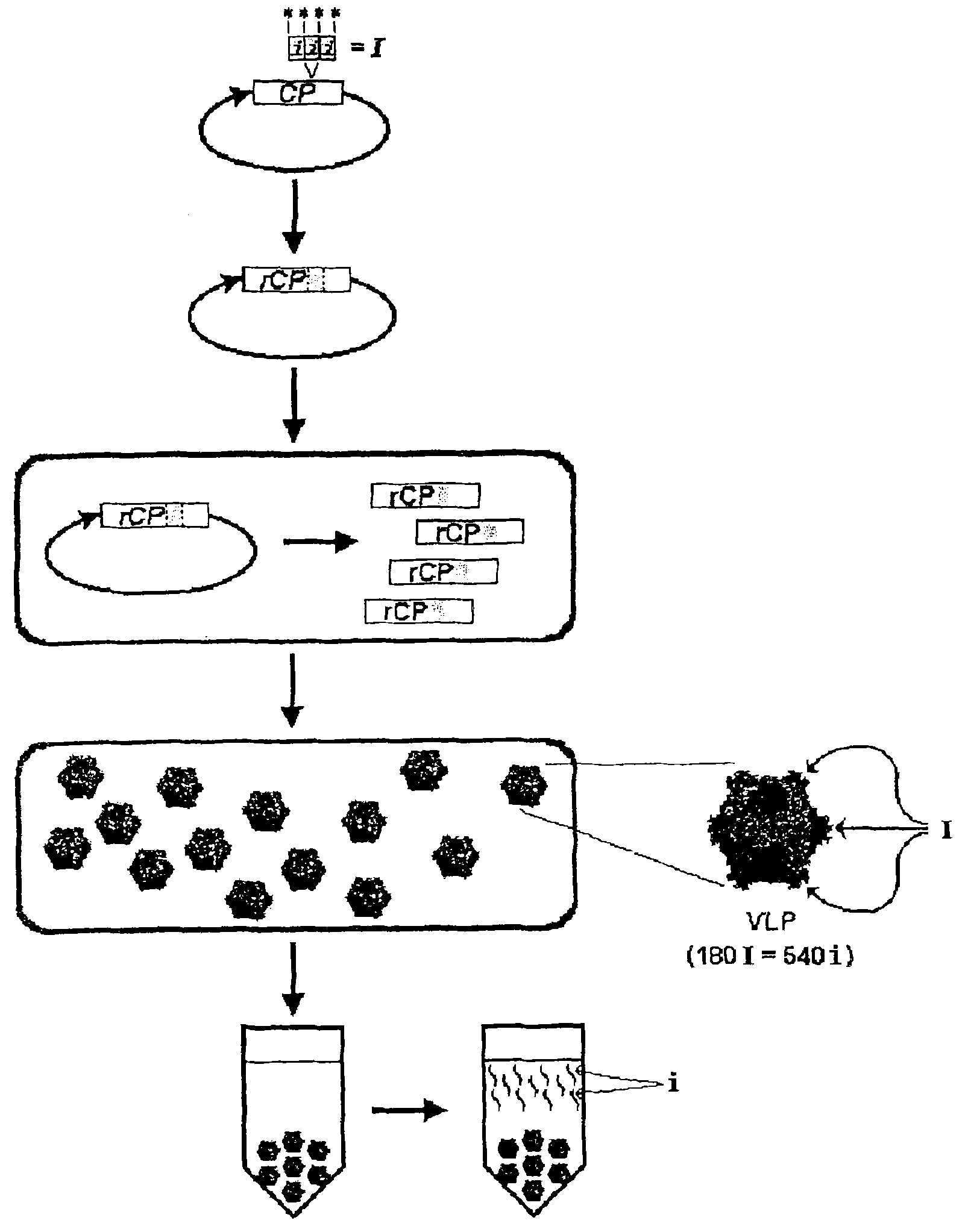
[0528] Example 3: Restriction digest based cloning and expression of influenza vaccine M2e peptide fused to the codon and hydrophilicity optimized 129 surface loop of the CCMV capsid protein
[0529] Peptide Synthesis:
[0530] Inserts were synthesized by overlapping DNA oligonucleotides described below and the thermal cycling procedure detailed below:
[0531]
[0532]
[0533] *(from Invitrogen Corp, Carlsbad, CA)
[0534] use PCR purification kit (Qiagen) purified PCR products, digested with XbaI (NEB), and The kit was purified again and ligated with T4 DNA ligase (NEB) into the XbaI-restricted CCMV CP Pseudomonas fluorescens expression vector (from Example 2) containing the XbaI restriction site in loop 129 point. After purification with a Micro Bio-spin 6 column (Biorad), the ligated product was transformed into Pseudomonas fluorescens strain DC454 (ΔpyrFRXF01414(lsc)::lacIq1) by electroporation. After shaking in LB medium at 30°C for 2 hours, the transforman...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com