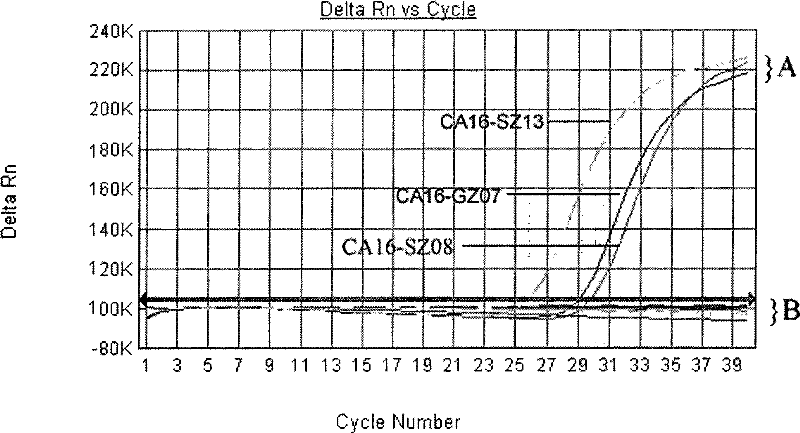
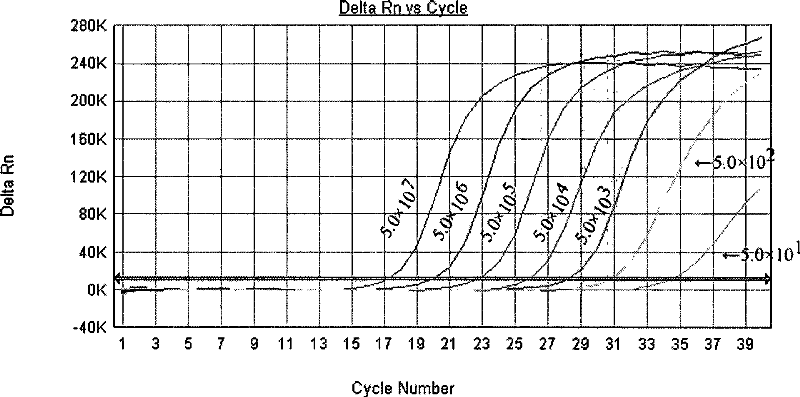
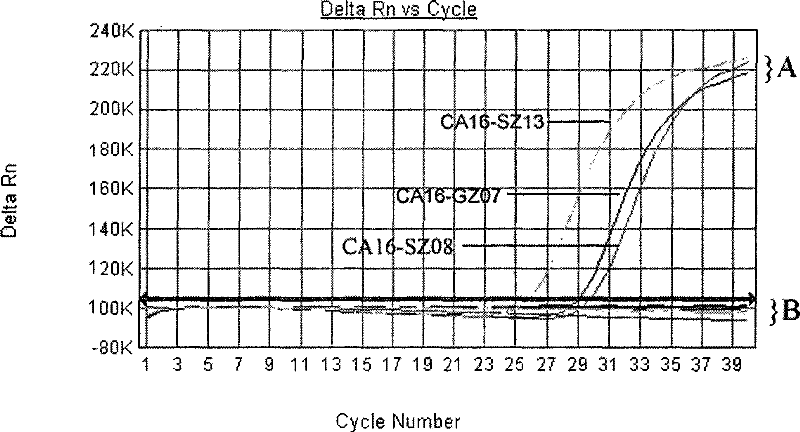
Primers, probes and kits for detection of coxsackie virus a16 nucleic acid
A Coxsackie virus, A16 technology, applied in the field of probes, kits, and primers for Coxsackie virus A16 nucleic acid detection, can solve the problems of cumbersome steps, long detection period, easy to miss detection, etc., and achieve high accuracy , high sensitivity, good primer specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] The following examples are used to further illustrate the present invention, but are not intended to limit the scope of the present invention.
[0021] Design and synthesis of primers and probes:
[0022] According to the CA16VP1 gene sequence in GenBank, primers and Taqman probes were designed at its conserved sites using the biological software Primer Express V2.0, and Blast homology comparison verification was carried out through the NCBI database. The primer sequences are:
[0023] CA16Pf (Forward): 5'-GAACCATCACTCCACACAGGAG-3'
[0024] CA16Pr (reverse): 5'-GTACCCGTGGTGGGCATTG-3'
[0025] The sequence of the probe is: 5'-CAGCCATTGGGAATTTCTTTAGCCGTG-3'
[0026] The 5' end of the probe was labeled with the reporter fluorescent dye VIC, and the 3' end was labeled with the quencher fluorescent dye BHQ2.
[0027] Extraction of RNA:
[0028] Extract RNA from virus samples with Roche High Pure viral RNA kit, take about 0.5 g of feces, add 500 μl of TE to make a 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com