Schistosoma japonicum microsatellite locus and application thereof
A technology of microsatellite loci and schistosomiasis, applied in the fields of biotechnology and genetics, can solve problems such as lack
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1 SSR acquisition and primer design
[0073] 1. DNA extraction of adult samples
[0074] Using DNA phenol extraction and ethanol precipitation method:
[0075] 1) Take a single insect and put it in 200μl extraction buffer to wash the insect body 3 times, and centrifuge at 2000g for 2min to separate the washing liquid;
[0076]2) Aspirate the washing solution, add 20μl extraction buffer, and use an RNAase-free sterile small grinding rod to grind the insect body to a homogenate;
[0077] 3) Add extraction buffer to a final volume of 50μl;
[0078] 4) Add 0.5μl RNAase (10ug / μl), 5μl SDS (10%), 0.5μl protein digestion enzyme, and mix with a gun repeatedly;
[0079] 5) Digestion in a compound water bath constant temperature oscillator at 110 rpm and 56°C for 1 hour;
[0080] 6) Add an equal volume of 50μl of a 25:24:1 mixture of phenol:chloroform:isopropanol, mix well, 10000g, and centrifuge for 5min;
[0081] 7) Take the supernatant, add 50μl chloroform: isopropanol 24:...
Embodiment 2
[0102] Example 2. Genetic analysis taking locus sjpn1 as an example
[0103] 1. Based on the Sjpn 1 sequence, design the corresponding primers as follows:
[0104] F: TGAGCACAACTGTATATCCCAAA
[0105] R: TGGGCAGACATACCAGGTTC
[0106] 2. PCR reaction, amplify the corresponding target fragment on the whole genomic DNA of the Schistosoma japonicum sample.
[0107] The PCR cycle conditions are: 95°C for 5 minutes; 94°C for 45s, 55°C for 45s, 72°C for 45s, 30 cycles; 72°C for 10 minutes. The PCR reaction system is: 25ng genomic DNA, 2 units of SBS Taq polymerase, two-way primers 10pm each, 1.25mM MgCl2, 1μl 10× reaction buffer, 0.5μl dNTPs (2.5mM, TaKaRa).
[0108] 3. The PCR products are purified by conventional methods.
[0109] 4. Analysis of genetic characteristics, such as analyzing multiple genetic characteristics (such as Tongling, Guichi, Duchang, Changde, Yueyang, Shashi, Xichang). The available genetic analysis methods are as follows:
[0110] (1). Electrophoresis identificati...
Embodiment 3
[0115] Example 3. Preliminary detection and gene scanning of PCR amplified products
[0116] 1. Preliminary detection of PCR amplification products
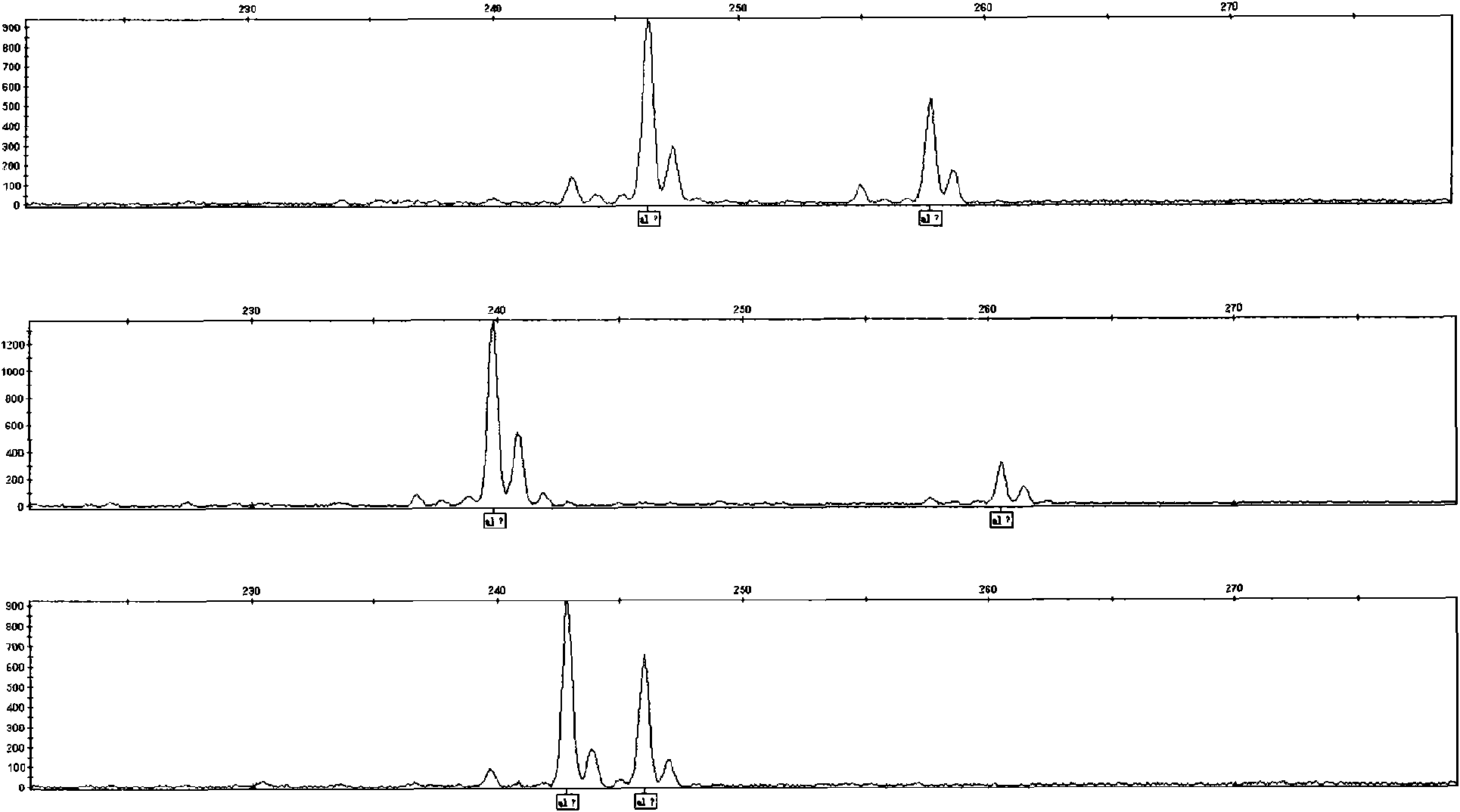
[0117] Before performing gene scanning (Genscan) on the PCR products, the amplified PCR products are detected to ensure the success of the gene scanning experiment. The amplified PCR product should have a positive target band, and the band type of the target band should be single without dragging. figure 1 The electropherograms of PCR amplification products at site sjpn8 of 96 individual samples in Wuhan epidemic area. A single band of interest appears in each lane, with a size of about 250 bp.
[0118] 2. Operation of GeneMapper 4.0
[0119] Set up different control panels (Panel) according to the size of PCR products amplified from different sites, and run GeneMapper 4.0, the operation can refer to the instruction manual.
[0120] 3. Genetypes
[0121] Since Schistosoma japonicum is a diploid organism, there are two genotypes in a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com