A microarray system and a process for producing microarrays
A technology of microarrays and microbeads, applied in the field of microarrays, can solve the problems of complex preparation of microarrays, time-consuming, complex and expensive instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
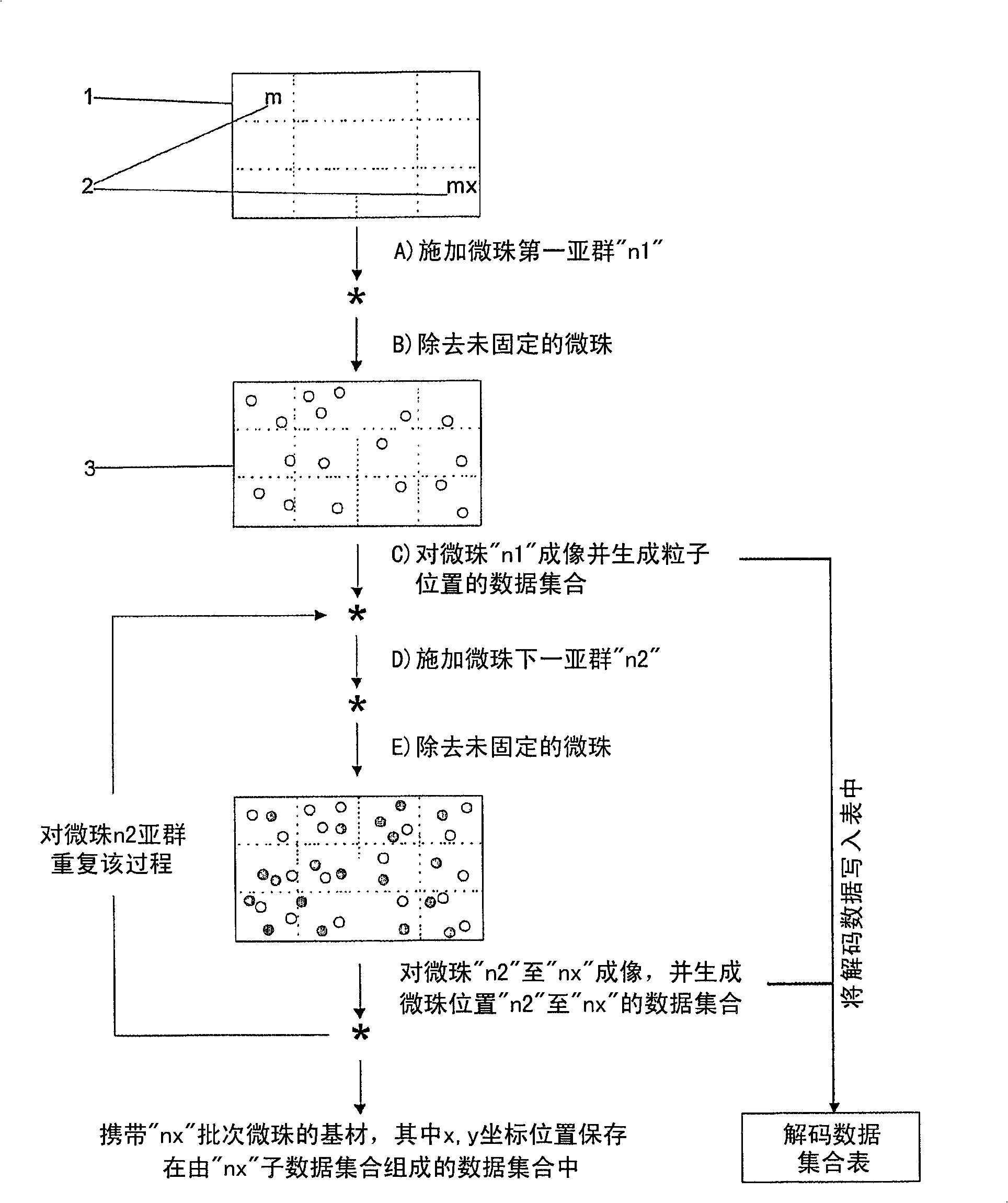
[0166] This example illustrates the use of bead-based microarrays for the detection of single nucleotide polymorphisms (SNPs).
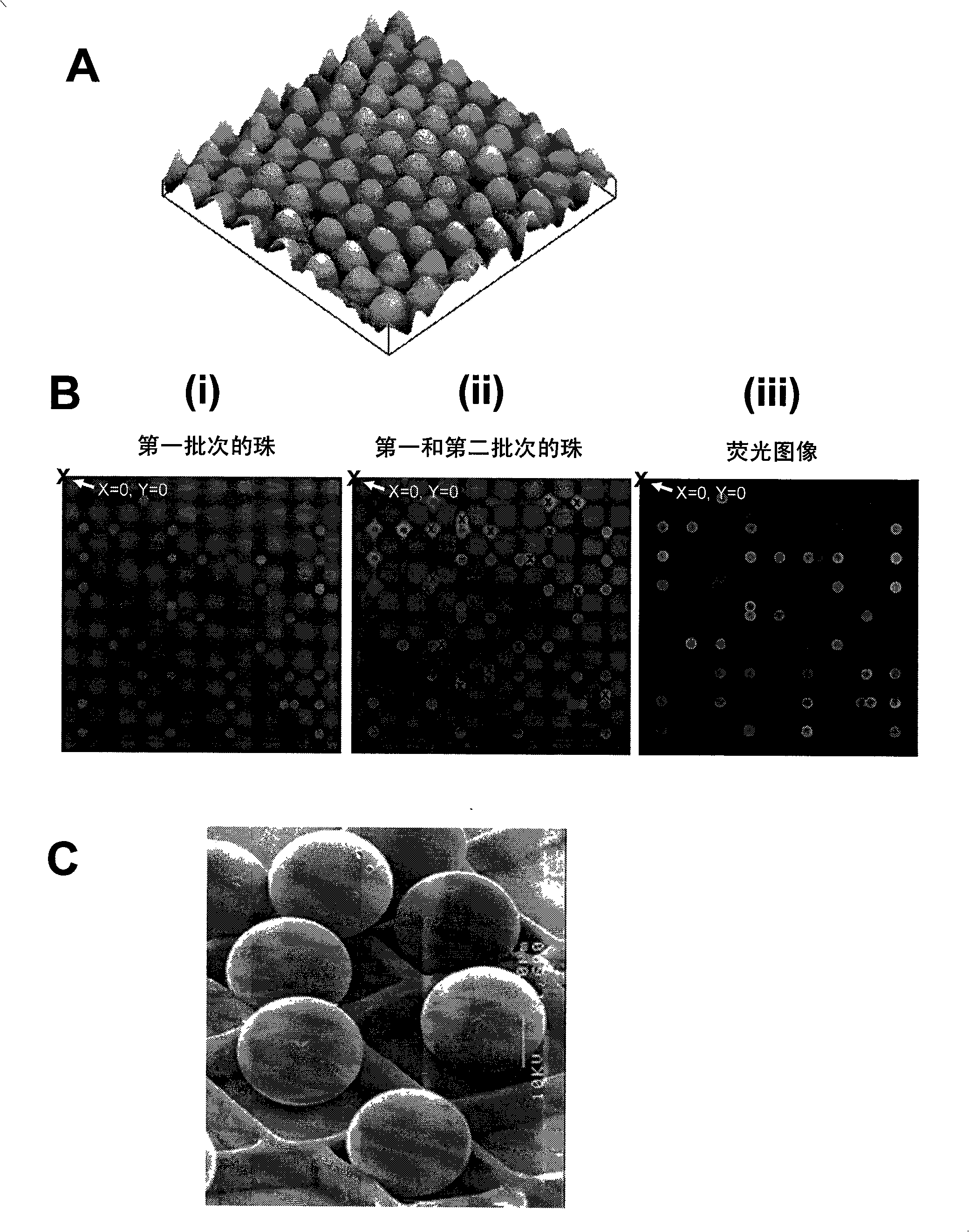
[0167] Arrays of polyacrylamide gel pads were prepared on the surface of glass slides using ultraviolet photopolymerization. Each discrete gel pad is further composed of a square array of micropillars. Four sets of beads were prepared by separately conjugating the beads with oligonucleotide probes targeting specific SNPs. These four probes contain all possible single nucleotide substitutions of the ATGC at the target SNP site. Using a robotic dispenser, immobilize the first set of conjugated beads on designated gel pads using microtargeting. Loosely immobilized beads were washed away and the remaining immobilized beads were imaged using a CCD camera system. The positions of all immobilized beads are decoded from this image, and the data is stored in computer memory. This process was then repeated sequentially for a second set of conjugated beads ...
Embodiment 2
[0170] This example illustrates the use of bead-based microarrays for the detection of fecal indicator bacteria in environmental samples.
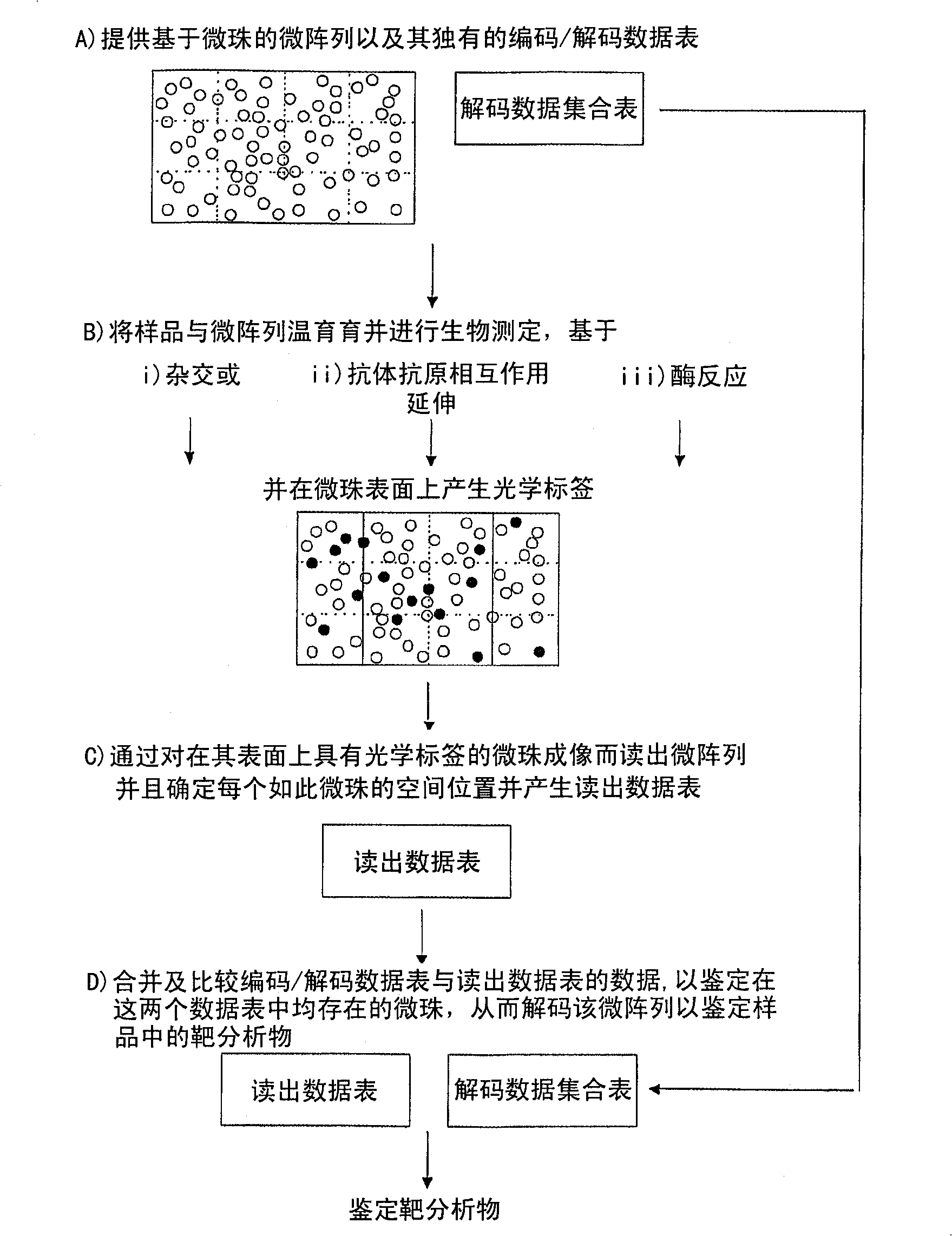
[0171] Twenty sets of probes were designed to target the 16S rRNA region of ten stool indicators. Each stool indicator is targeted by a pair of perfect match (PM) probes and a mismatch (MM) probe that differs by one single nucleotide in the central portion. Using the same method described in Example 1, ten sets of beads conjugated to PM probes were immobilized on different gel pads, and the position of the beads in each gel pad was characterized. This step is repeated for beads conjugated to MM probes, so that each gel pad contains beads conjugated to pairs of probes targeting a specific fecal indicator. These two groups of beads are distinguished based on their spatial codes.
[0172] Environmental samples suspected of containing fecal indicators were prepared and hybridized to immobilized beads. The presence of fecal indicators in unk...
Embodiment 3
[0174] This example demonstrates the performance of antibody-based assays using antibody-antigen interactions and DNA-based assays using DNA / DNA or DNA / RNA hybridization in parallel on the same chip.
[0175] Antibody preparation by biotinylation :
[0176] Antibodies (polyclonal mouse IgG and polyclonal mouse anti-IgG) were diluted to approximately 2 mg / ml. Biotin-NHS (10 mg / ml, dissolved in DMSO) was added to the antibody solution at a molar ratio of 1:1, 5:1 and 10:1 (M biotin:M antibody), and the reaction tube was placed at 25 Incubate overnight at °C. The biotinylated antibody was purified using a gel filtration column (D-salt dextran desalting column, Pierce.). Biotinylated antibodies were dissolved in PBS and stored at -20°C.
[0177] Method for FITC conjugation of antibodies :
[0178] Antibodies (polyclonal mouse IgG and polyclonal mouse anti-IgG, Arista Biological inc.) were diluted to approximately 2 mg / ml. FITC (10 mg / ml, dissolved in DMSO) was added to th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com