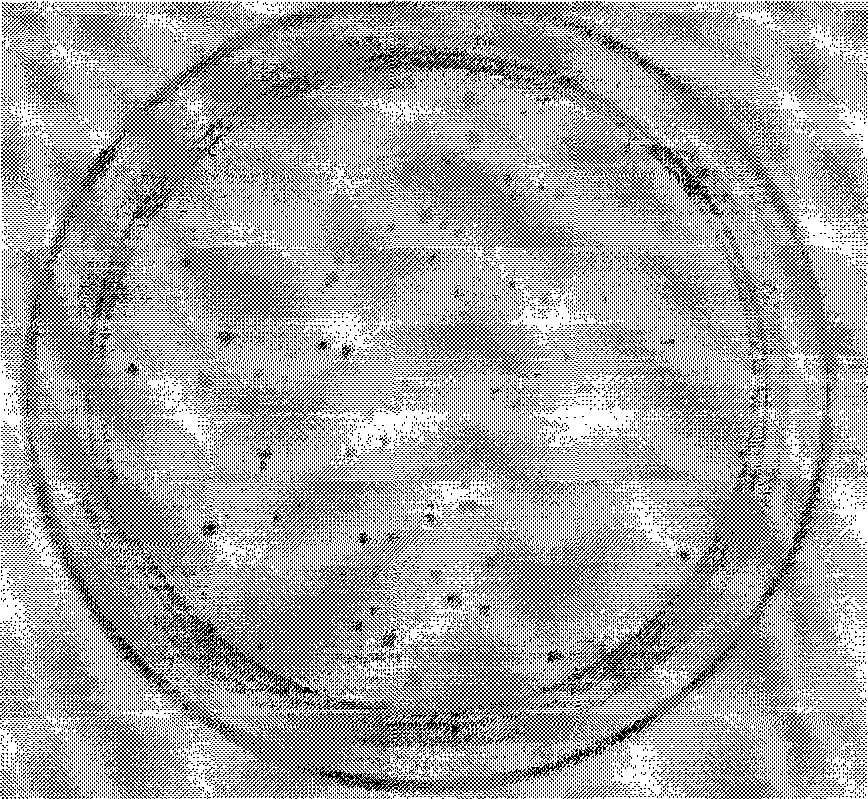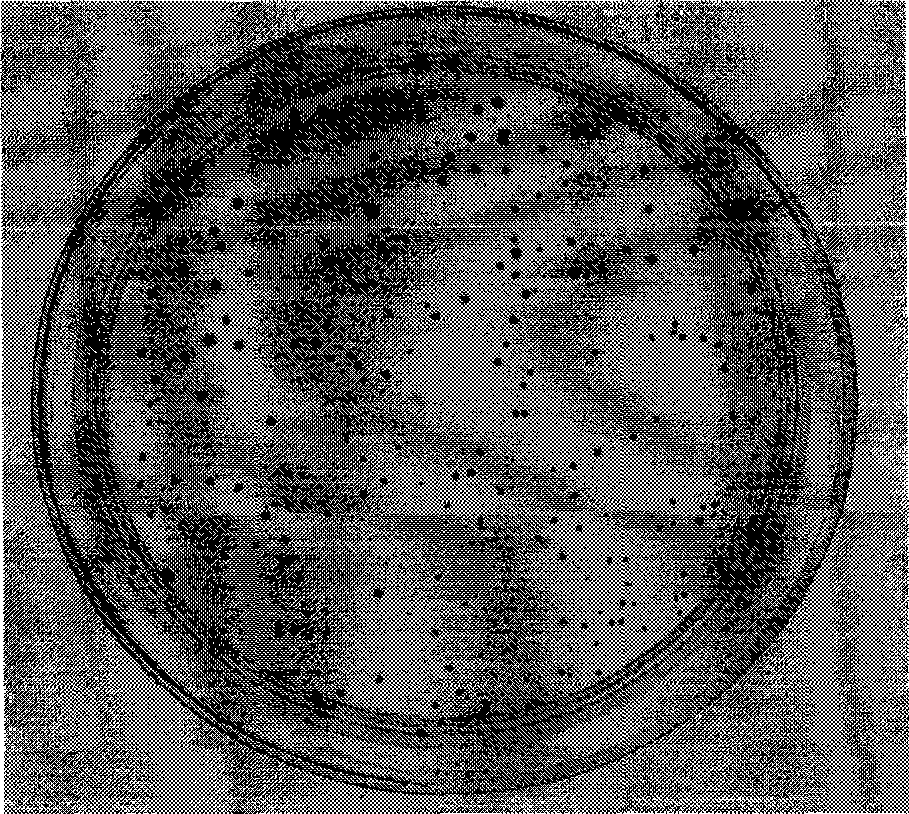Phi C31 site specific recombinase mutant, preparation method thereof and application
A mutant and recombinase technology, applied in the field of genetic engineering, can solve the problem of low efficiency of mediating site-specific integration, and achieve the effect of improving recombination activity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Verification of the activity of ΦC31 site-specific recombinase mutants on prokaryotic cells
[0019] After loading the ΦC31 site-specific recombinase mutant gene sequence into the pINT-T expression vector, transform the E.coli cells carrying the pRES-MpsL1 vector, and spread it on the X-gal plate. After induction at different temperatures (30°C to 37°C first), observe the appearance time (3-12h), color depth and quantity of blue clones.
[0020] The result is as figure 1 with figure 2 As shown, 4 hours after temperature induction, almost all clones on the plate expressing the ΦC31 site-specific recombinase mutant were blue, and the blue color was darker, while the plate with the ΦC31 site-specific recombinase as the control On , there are fewer blue clones and they are lighter in color. This shows that the ΦC31 site-specific recombinase mutant has higher activity.
Embodiment 2
[0021] Example 2 Verification of the activity of ΦC31 site-specific recombinase mutants on eukaryotic cells
[0022] Construct eukaryotic expression vectors of ΦC31 site-specific recombinase mutants and eukaryotic expression vectors carrying attB sites, transfect mouse 3T3 cells, and apply 200 μg / ml hygromycin B to 30% of transfected cells 48 hours later. After 14 days of drug screening, clones were counted; single clones were amplified, and DNA was extracted by DNEasy; the recombination results were detected by PCR. The sequencing results of the PCR product showed that the first 10 bp of the obtained sequence was the attB site sequence, and the exchange center TTG (nt.11-13) was followed by the mouse genome mpsL1 site sequence. This shows that the ΦC31 site-specific recombinase mutant of the present invention can specifically mediate gene recombination at the mpsL1 site.
Embodiment 3
[0023] Example 3 Verification of integration efficiency of ΦC31 site-specific recombinase mutants on eukaryotic cells
[0024] 50ng of plasmid pHZ-attB and 5μg of expression mutant plasmids were co-transfected into mouse 3T3 cells, and the plasmid pCmv-Int of φC31 integrase was used as a control, and the Lipofectamine method was used to complete. 48 hours after transfection, 70% of the transfected cells were subjected to tissue DNA extraction. Genomic DNA was extracted with the DNEasy tissue kit. These DNAs were used for quantitative PCR detection to find the frequency of recombination at the mpsL1 locus. Design PCR forward primers and reverse primers based on the attB sequence and mpsL1 site sequence, and design a probe attB sequence integrated in the mpsL1 site binding region, which is mainly used to detect site-specific reactions at the mpsL1 site. Quantitative detection of PCR was accomplished by SYBR Green kit (Finzymes, F-430S), and the amplification reaction was carri...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com