Typing method for detection of HCMV gBn by real-time fluorescent quantitative PCR
A technology of real-time fluorescence quantification and typing method, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc. It can solve the problems of inability to quantify DNA, etc., and achieve the effect of avoiding human judgment, intuitive results, and a wide range of quantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Equipment: PCR instrument SLAN (Shanghai, Hongshi), UV spectrophotometer HP8453 (HP company, Germany), gel image analysis system BioSenSC300 (Shanghai Shanfu Scientific Instrument Co., Ltd.), low-temperature high-speed centrifuge (Biofuge 28RS) (Heraeus company, Germany), constant temperature water bath box (Model DK-8D (Shanghai Senxin Experimental Instrument Co., Ltd.).
[0036] Reagents: TAQ enzyme (Promega, USA), nucleic acid extract (Shanghai Zhijiang Biology), primers (Invitrogen, USA). Specific ratio of nucleic acid lysate: 10mmol / L Tris-cl (pH8.0), 0.1mol / L EDTA (pH8.0), 20ug / ml Trypsin, 0.5% SDS.
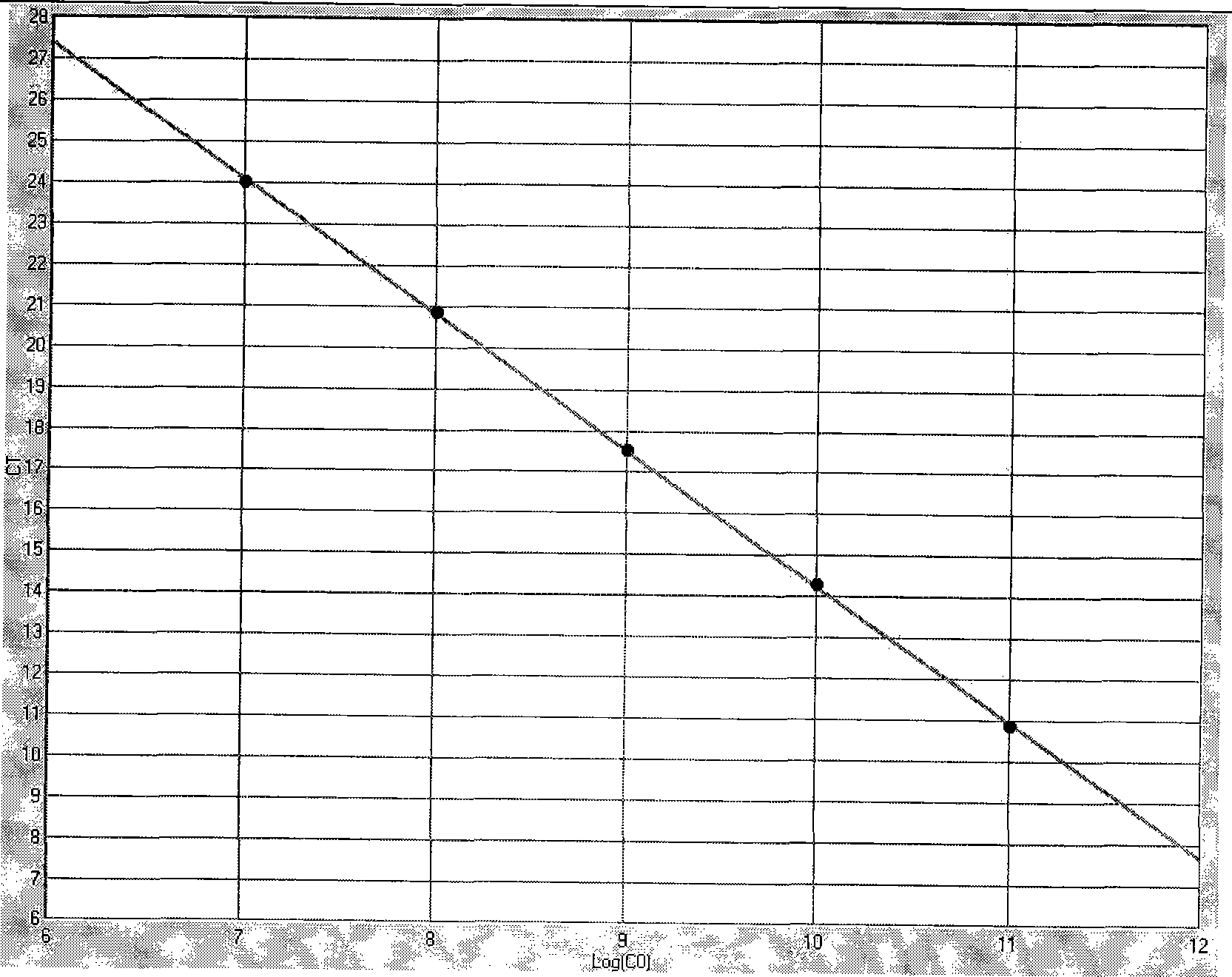
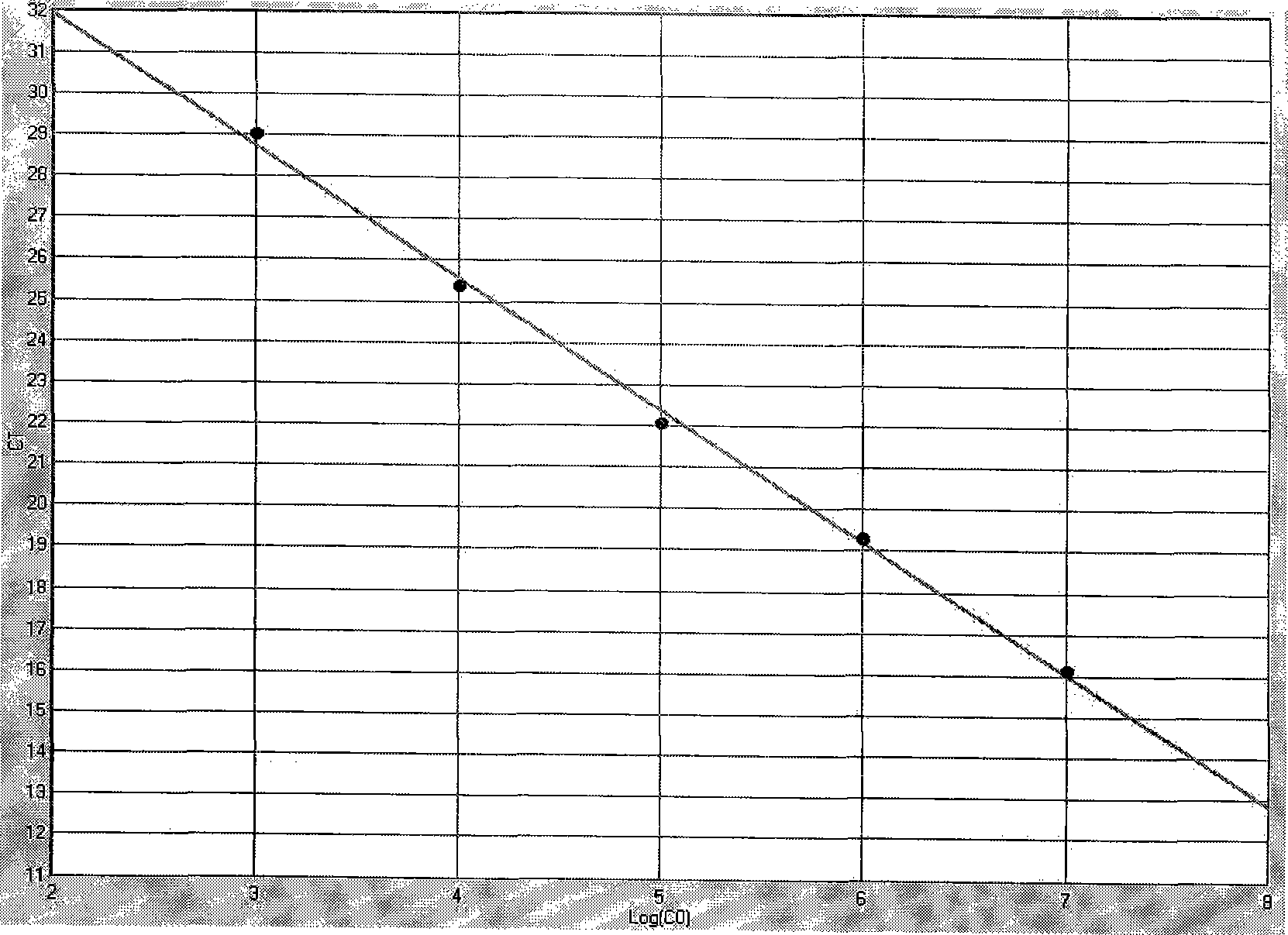
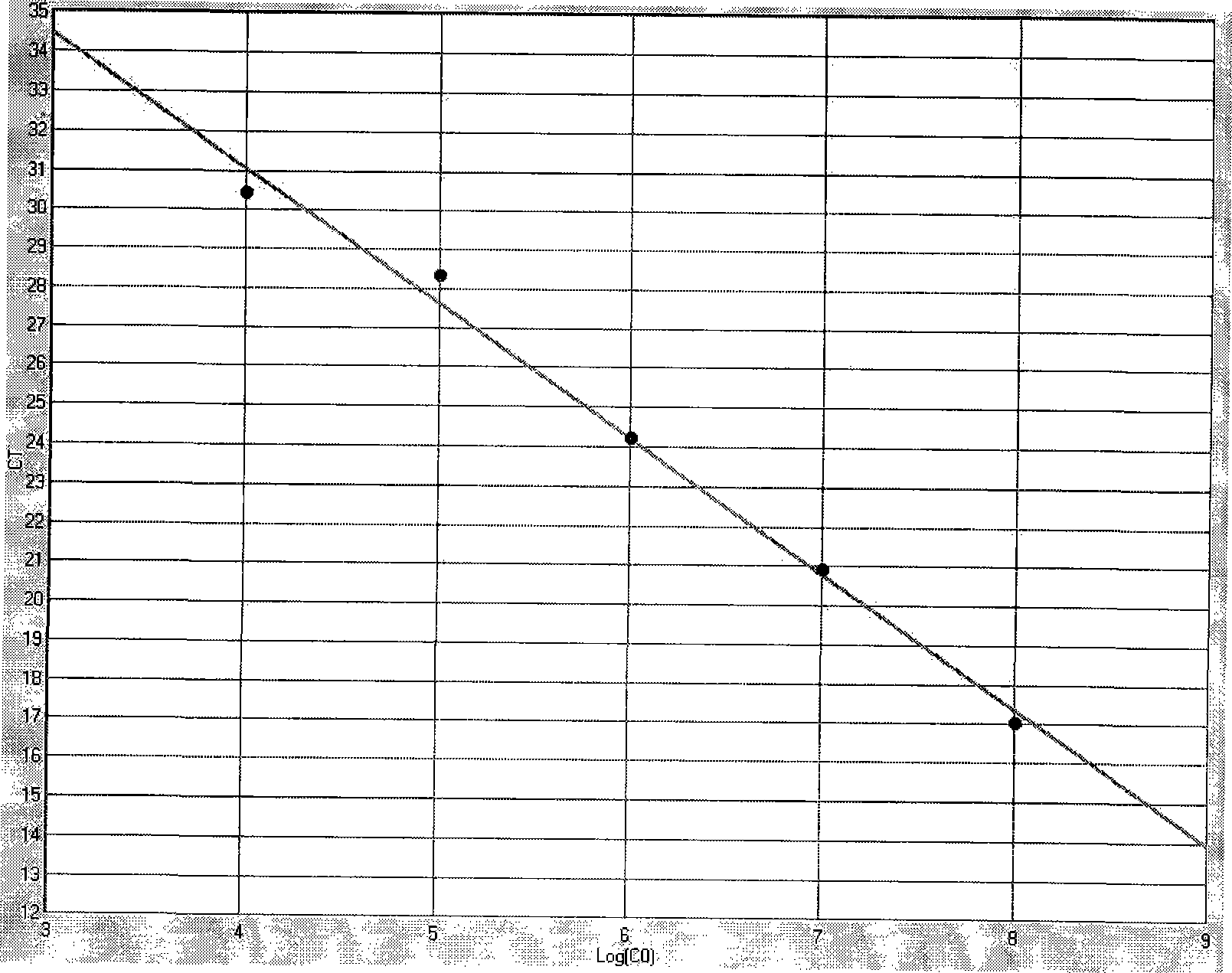
[0037] PCR Primers and Probes
[0038] HCMV-gB1-1:5'-cggttcagtctctcaacg-3'
[0039] HCMV-gB1-2: 5'-caccacatctccgtacttg-3'
[0040] Probe HCMV-gB1-3: FAM-ttcccaaacggtcagc-BHQ1-3;
[0041] HCMV-gB2-1: 5'-ctcgtacgacgtcgtctcaa-3'
[0042] HCMV-gB2-2: 5'-cacacgcgataggggtactt-3'
[0043] Probe HCMV-gB2-3: 5'-FAM-gagaatagactgac-BHQ1-3';
[0044] HCMV-gB3-1: 5'-ctcgta...
Embodiment 2
[0077] According to the above experimental procedures, 1207 samples were selected for real-time fluorescence quantitative PCR to determine the genotypes of HCMVgBn1, 2, and 3 after transplantation recipients, including 40 liver transplant recipients with 366 specimens, and 120 kidney transplant recipients with 350 specimens , 55 cases of bone marrow transplant recipients, 481 samples, the results showed that the positive rate of gBn1 type was 5.3%, the positive rate of type 2 was 1.0%, the positive rate of type 3 was 2.7%, and type 4 was not detected; The difference (p>0.05), the mixed infection of gBn1 and type 3 accounted for 42.3% of gBn1 positive; the distribution of different transplant types in the positive samples had no difference. Due to the quantification of HCMV gB, it can not only detect the infection status of HCMV, but also judge the prognosis of patients. For example, the prognosis of type 1 infection is generally better; mixed infection has higher viral load and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com