Method for preparing nucleic acid molecular weight label
A molecular weight and nucleic acid technology, applied in the field of molecular biology, can solve problems such as low production efficiency and cumbersome steps of nucleic acid molecular weight marking
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
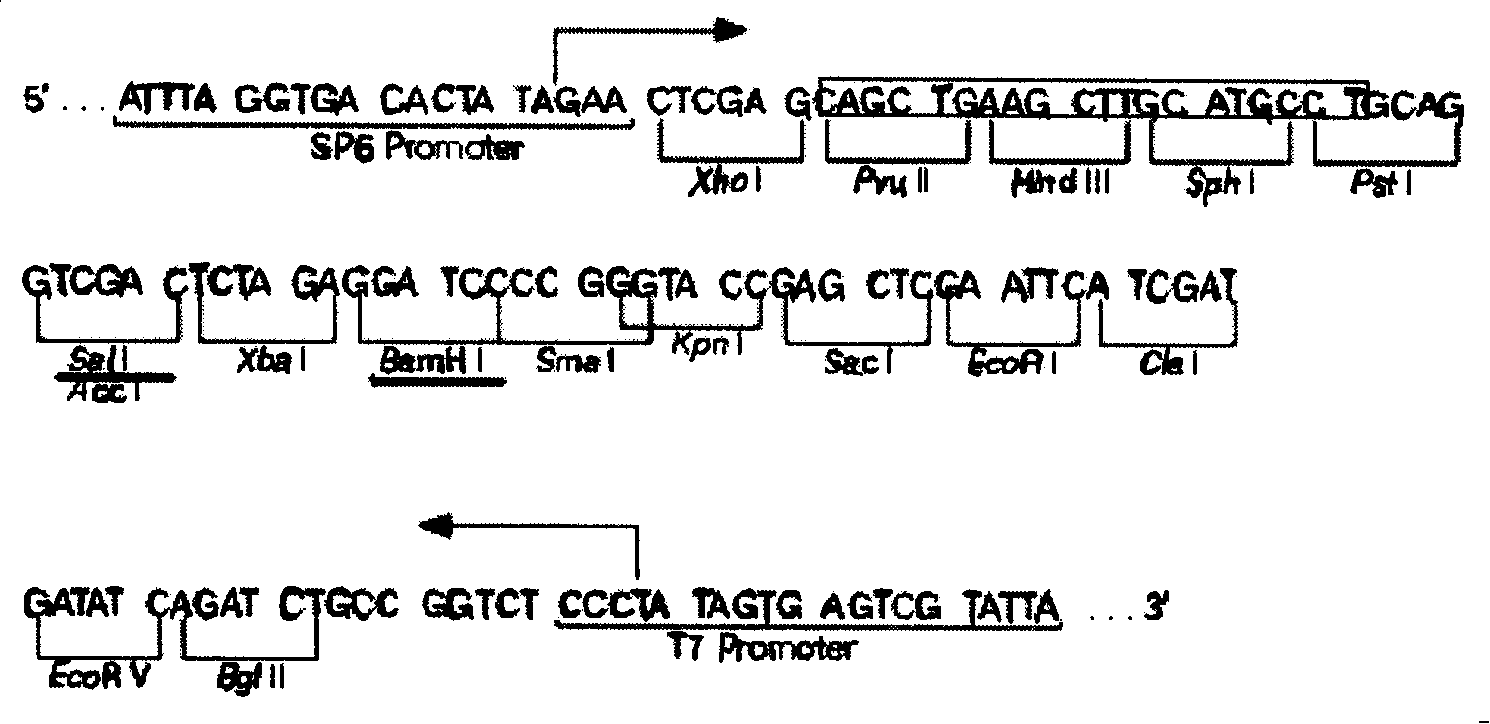
[0033] figure 1 It is the multiple cloning restriction site of the psp72 plasmid. In this experiment, two restriction sites, Sal I and BamHI, were selected, and the vector after digestion was recovered for 1.5 hours to obtain the double digestion vector.
[0034] The synthesized sequence is a single-stranded oligonucleotide, which is synthesized by the company according to the designed sequence (Shanghai Sangon Bioengineering Technology Service Co., Ltd., Lot No: AA26474, AA26475). The two strands are complementary. Anneal to form double strands first. Annealing conditions are as follows:
[0035] Nuclease-free H 2 O 40ul
[0036] 5×annealing buffer 20ul
[0037] DNA oligoA 20ul
[0038] DNA oligoB 20ul
[0039]
[0040] 100ul
[0041] 95°C×2min, drop 0.5°C every 40 seconds, and store at 4°C after dropping to 25°C (annealing buffer is a product of Biyuntian Company)
[0042] as figure 2 As shown, the bold it...
Embodiment 2
[0050] Figure 4 As shown, inserts of different lengths are used to make markers, and several bases can be randomly added to the original insert. In this embodiment, 50 bases (lowercase letters) are added on the original basis to make the primers The length of the sequence between them reaches 150bp, while the original enzyme cutting site and primer binding site remain unchanged. It can be seen that markers of various molecular weights can be produced by designing and synthesizing insert sequences of different lengths, saving time and effort.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com