Method for producing purine nucleosides and nucleotides by fermentation using bacterium belonging to the genus bacillus or escherichia
A purine nucleotide and purine nucleoside technology, applied in the field of purine nucleoside production, can solve the problems of no Bacillus amyloliquefaciens, no effect of YdhL, and no effect yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1. Cloning of the Bacillus subtilis ydhL gene
[0100] The entire nucleotide sequence of the Bacillus subtilis subsp. 168 strain has been determined (Kunst et al., Nature 390, 249-256 (1997)). Based on the reported nucleotide sequence, primers ydhL_N (SEQ ID No. 5) and ydhL_C (SEQ ID NO: 6) were synthesized for PCR amplification of the ydhL gene from Bacillus subtilis strain 168. Primer ydhL_N is consistent with the sequence of 489 to 467 bp upstream of the start codon of ydhL gene, and has a restriction enzyme SalI recognition site introduced at its 5'-end. Primer ydhL_C is complementary to the 58 to 80 bp sequence downstream of the stop codon of ydhL gene, and has a restriction enzyme BglII recognition site introduced at its 5'-end.
[0101] Chromosomal DNA of the Bacillus subtilis 168 strain was prepared by the usual method. PCR was carried out on "Perkin Elmer GeneAmp PCR System 2400" according to the following conditions: 94°C for 30 seconds, 61°C for 45 ...
Embodiment 2
[0114] Embodiment 2. Cloning of Bacillus amyloliquefaciens ydhL gene
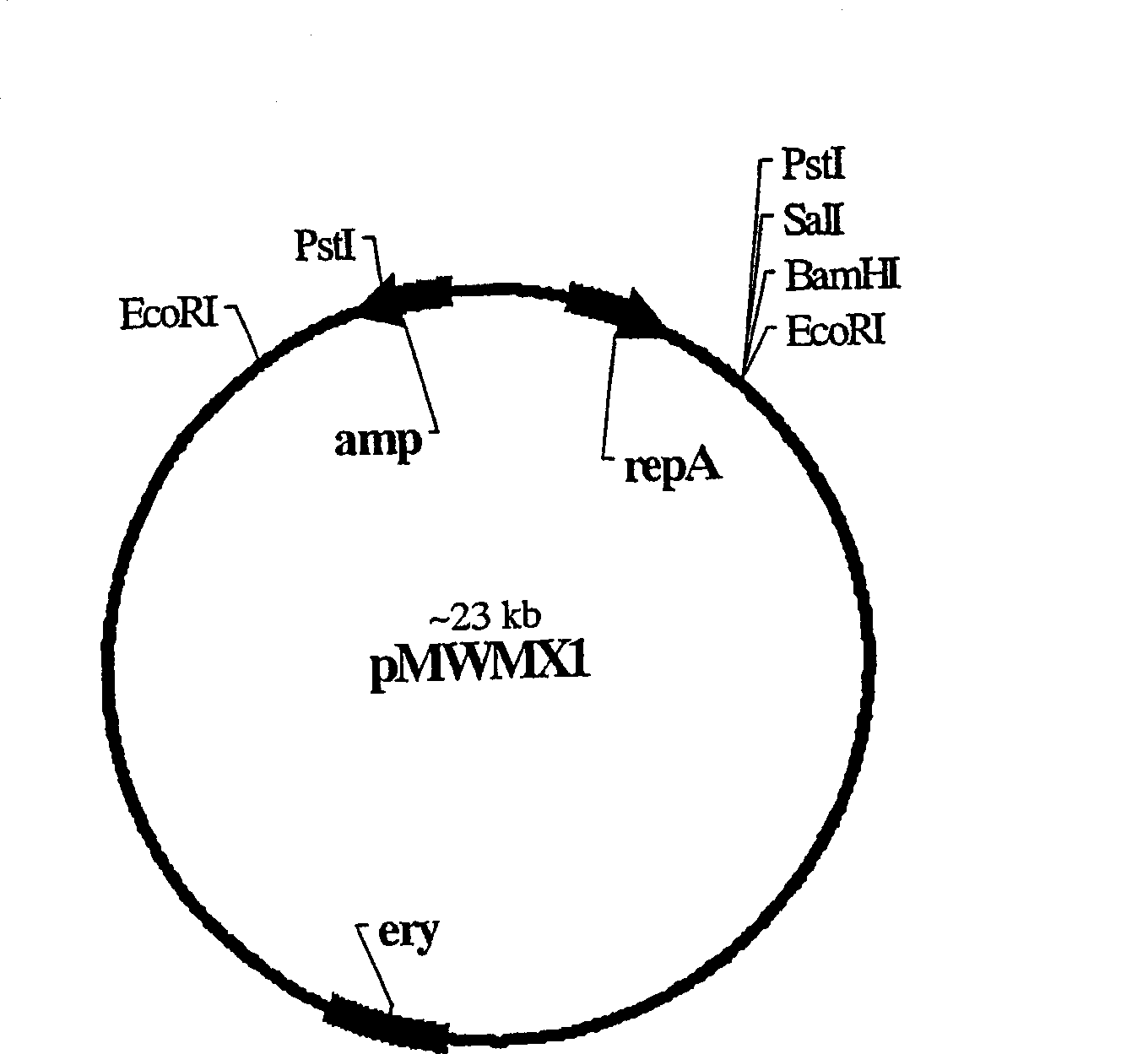
[0115] The data shown in Example 1 show that when simultaneously introduced into M9 glucose minimal agar (Table 1), the Bacillus subtilis ydhL gene (hereinafter referred to as ydhL Bs ) confers resistance to 2,6-diaminopurine and 6-mercaptopurine in cells. Based on this fact, the clone from Bacillus amyloliquefaciens ydhL Bs The orthologue of the gene (hereinafter referred to as ydhL Ba ). Chromosomal DNA of B. amylobacter strain BA1 (IAM1523) was prepared by the general method. The chromosomal DNA was cut with EcoRI enzyme and ligated with the pMW118 vector which had been treated with the same enzyme before. The resulting DNA was used to transform E. coli strain TG1. Transformants were selected on minimal agar containing 400 μg / ml 2,6-diaminopurine, 400 μg / ml 6-mercaptopurine and 100 μg / ml ampicillin. Plasmid DNA was isolated from transformants and analyzed. Thus, a hybrid plasmid pYDHL2 containing ...
Embodiment 3
[0116] Example 3.ydhL Bs and ydhL Ba Effect of Gene Amplification on Inosine Production of Escherichia coli Inosine-producing Strains
[0117] The inosine-producing E. coli strain FADRaddedd (pMWKQ) was transformed with the pMW18 vector, and the YDHL1 and pYDHL2 plasmids, respectively. Transformants were selected on L-agar containing 100 mg / l ampicillin and 75 mg / l kanamycin. Strains FADRaddedd (pMWKQ, pMW18), FADRaddedd (pMWKQ, pYDHL1) and FADRaddedd (pMWKQ, pYDHL2) were thus obtained. These strains were cultured in L-broth containing 100 mg / l ampicillin and 75 mg / l kanamycin, respectively, at 37°C for 18 hours, and 0.3 ml of the resulting culture was inoculated into 3 ml containing 100 mg / l ampicillin and 75 mg / l kanamycin. 75mg / l kanamycin in the fermentation medium, the fermentation medium was placed in a 20x200mm test tube, and cultured at 37° C. for 72 hours with a rotary shaker.
[0118] Composition of fermentation medium (g / l):
[0119] Glucose 40.0
[0120] (NH 4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com