Cell model for quick screening of histone deacetylase inhibitor
A deacetylase and inhibitor technology, which is applied in the field of cell models for rapid screening of histone deacetylase inhibitors, and can solve problems such as unfavorable gene transcription and gene expression shutdown.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
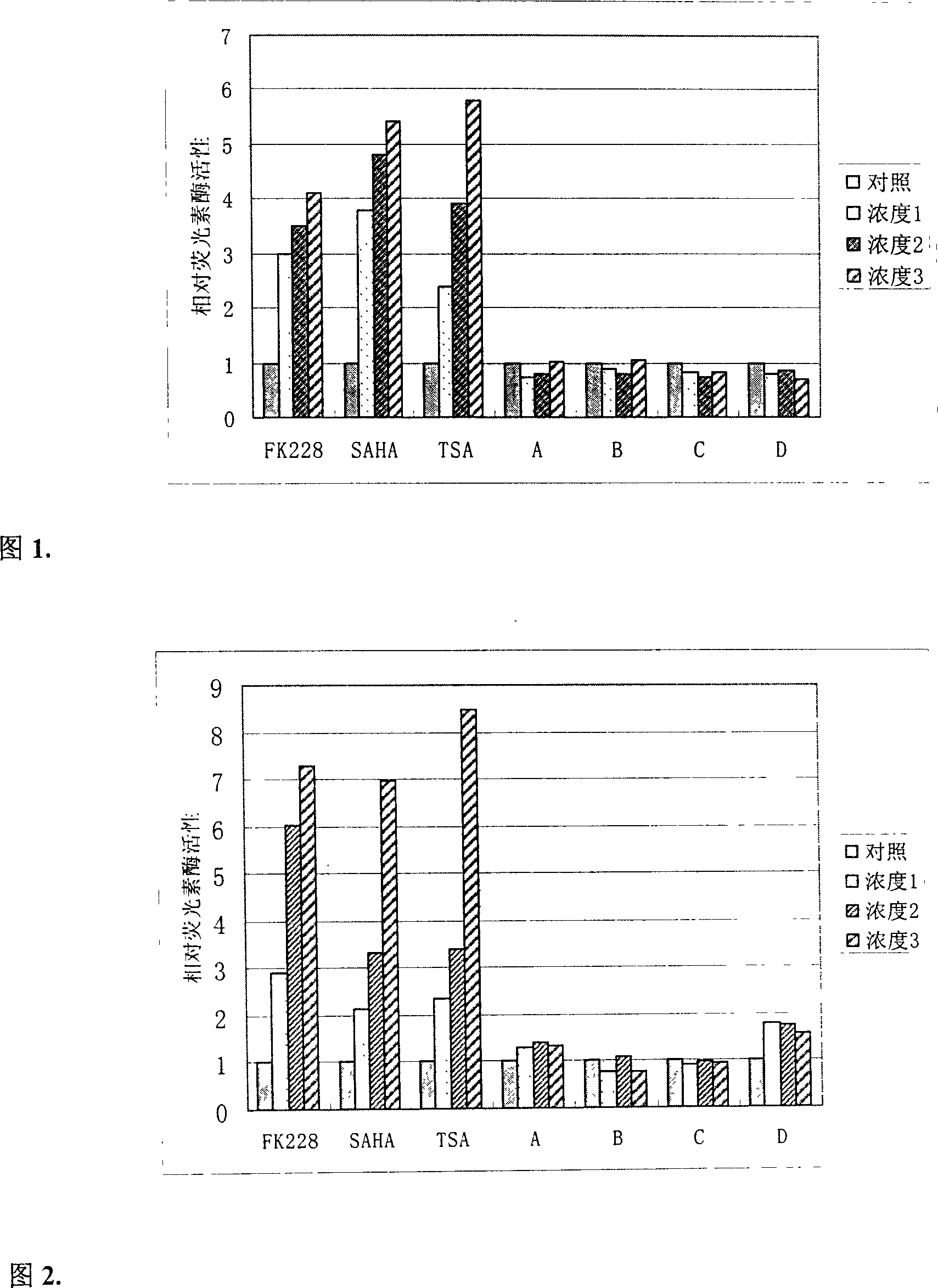
[0020] The following combination of different types of HDAC inhibitors Depsipeptide (FK228), suberoylanilidehydroxamic acid (SAHA), Trichostatin A (TSA) and other 4 kinds of drugs with anti-tumor activity 5-aza-2'-deoxycytidine (DAC) , retinoic acid, paclitaxel and 17-AAG respectively treated the experiment of the cell model to further illustrate the present invention.
[0021] 1. Experimental method
[0022] 1.1 Using the method of molecular cloning, the above-mentioned promoter sequence fragments were respectively inserted into the commercial vectors containing the luciferase reporter gene. After plasmid amplification and purification, the COS7 cells were transfected with liposome-mediated method, G418 (1mg / ml ) After pressurized selection for 21 days, a cell-resistant clone stably expressing the foreign gene was obtained, and the resistant cell could be used as a model for subsequent drug screening.
[0023] 1.2 HDAC inhibitors FK228 and TSA were provided by Dr. David Schr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com