Preparation method of oligonucleotide chip for detecting transplanted kidney function
A technology of oligonucleotides and oligonucleotide probes, applied in the field of oligonucleotide chips for detecting transplanted kidney function, can solve the problems of high price, heavy workload, and failure to detect, so as to ensure sensitivity and specificity speed, save time and money, and reduce disruptive effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1 Preparation and hybridization of the oligonucleotide chip of the present invention
[0019] 1. Chip Preparation
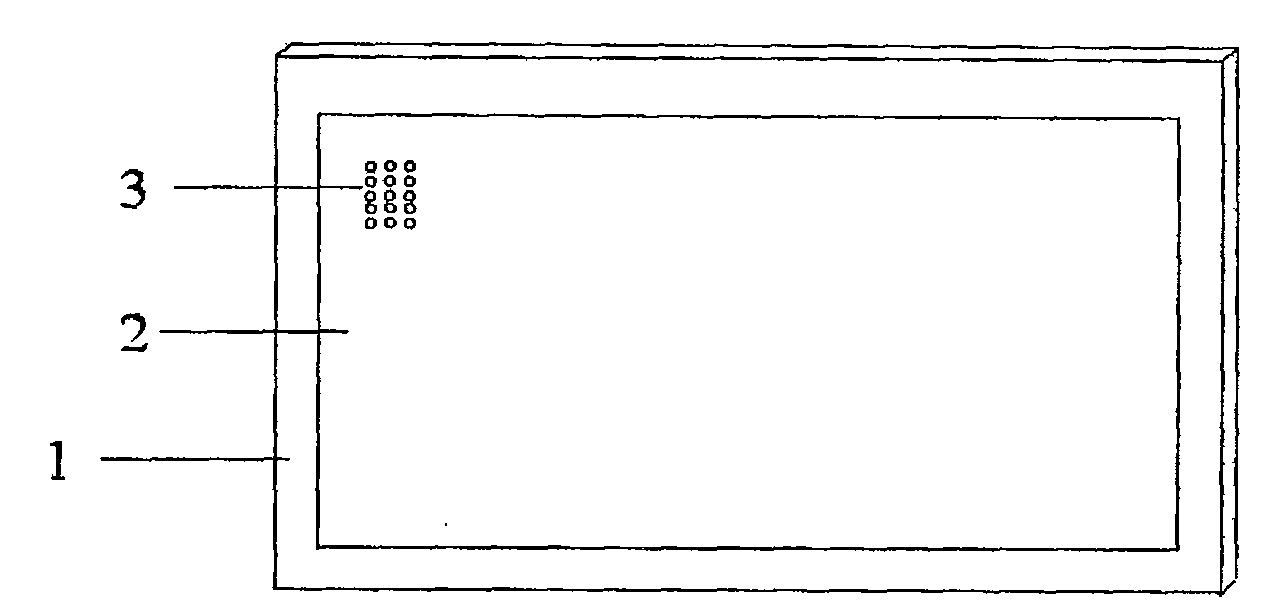
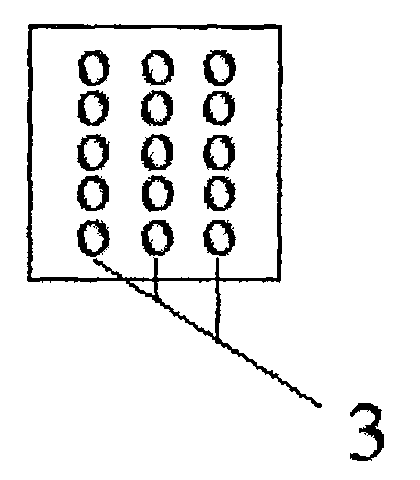
[0020] see figure 1 , the oligonucleotide chip is composed of a matrix nylon membrane 1, a sample area 2, and a modified and hybridized oligonucleotide probe 3 fixed on the surface of the nylon membrane and distributed in a matrix, and the oligonucleotide probe 3 has SEQ ID NO 1 the sequence of. Specific oligonucleotide probes 3 are respectively spotted on the spotting area 2 of the chip, and the spots are formed into 96 (12×8) matrices, each matrix includes 15 (5×3) spots, and the diameter of the spot is 0.6mm , with a dot pitch of 1.5 μm. According to the existing relevant knowledge, 449 immune-related genes were screened out and synthesized by BIO BASIC INC (Canda) using the 70 bp long oligonucleotide sequence provided by Operon Company, see the sequence of SEQ ID NO 1. The oligonucleotide synthesized at 1OD was dissolved in 100ul double ...
Embodiment 2
[0024] Embodiment 2 The application of the oligonucleotide chip prepared by the present invention
[0025] 1: The repeatability and accuracy of the chip
[0026] Pre-experiment 10 probes, the results of two independent hybridizations for each probe, R 2 The value is 0.88-0.95, according to the correlation coefficient R of the chip-to-chip data 2 >0.81 is recognized as the standard of good repeatability of the experiment, and the hybridization data obtained in this paper has good repeatability and can be used for subsequent analysis.
[0027] Compared with the acute rejection group and the stable renal function group, the 10 most significantly differentially expressed genes were screened out. The results of real-time quantitative PCR verification were consistent with the detection results of the microarray. They all showed high expression in the acute rejection group, which verified the microarray hybridization results. accuracy.
[0028] 2: Detection of clinical specimens ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com