Method for protein gel internal enzymolysis
A protein and trypsin technology, applied in the field of enzymatic hydrolysis of protein gel, can solve the problems of interfering with the effective ionization process of protein samples, unsatisfactory database search results, and insufficient enzymatic hydrolysis, etc., to achieve reliable protein identification results and improve Sequence coverage, the effect of improving enzymatic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
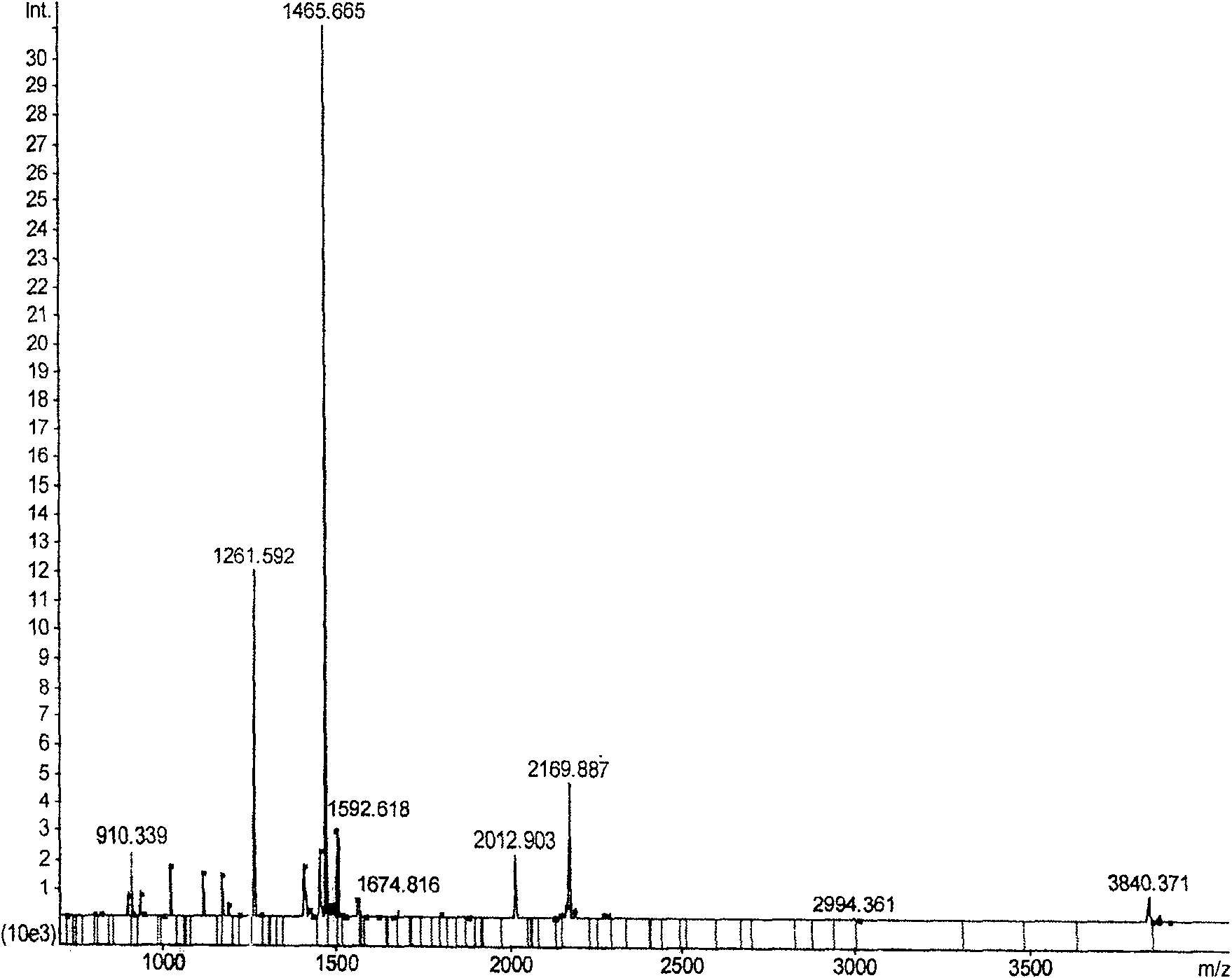
[0044] Example 1. Simplified in-gel enzymatic hydrolysis method to enzymatically hydrolyze Salicornia rubisco protein and its effect verification
[0045] In green plants, the content of Rubisco protein (ribμlose-1, 5-bisphosphatecarboxylase / oxygenase) is very abundant, which can reach more than 50% of the total soluble protein, and the Rubisco protein of different plants has high conservation (Rossignol M, Peltier J B , Mock H P, Matros A, Maldonado A M, Jorrín J V. Plant proteome analysis: A 2004-2006 update. Proteomics, 2006, 6: 5529-5548). In mass spectrometry research, Rubisco protein is usually used to test whether the relevant experimental conditions are suitable.
[0046] 1. Protein electrophoresis and extraction of target protein spots
[0047] According to Saravanan and Rose's improved phenol extraction method (Saravanan R S and Rose J C.A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics, 2...
Embodiment 2
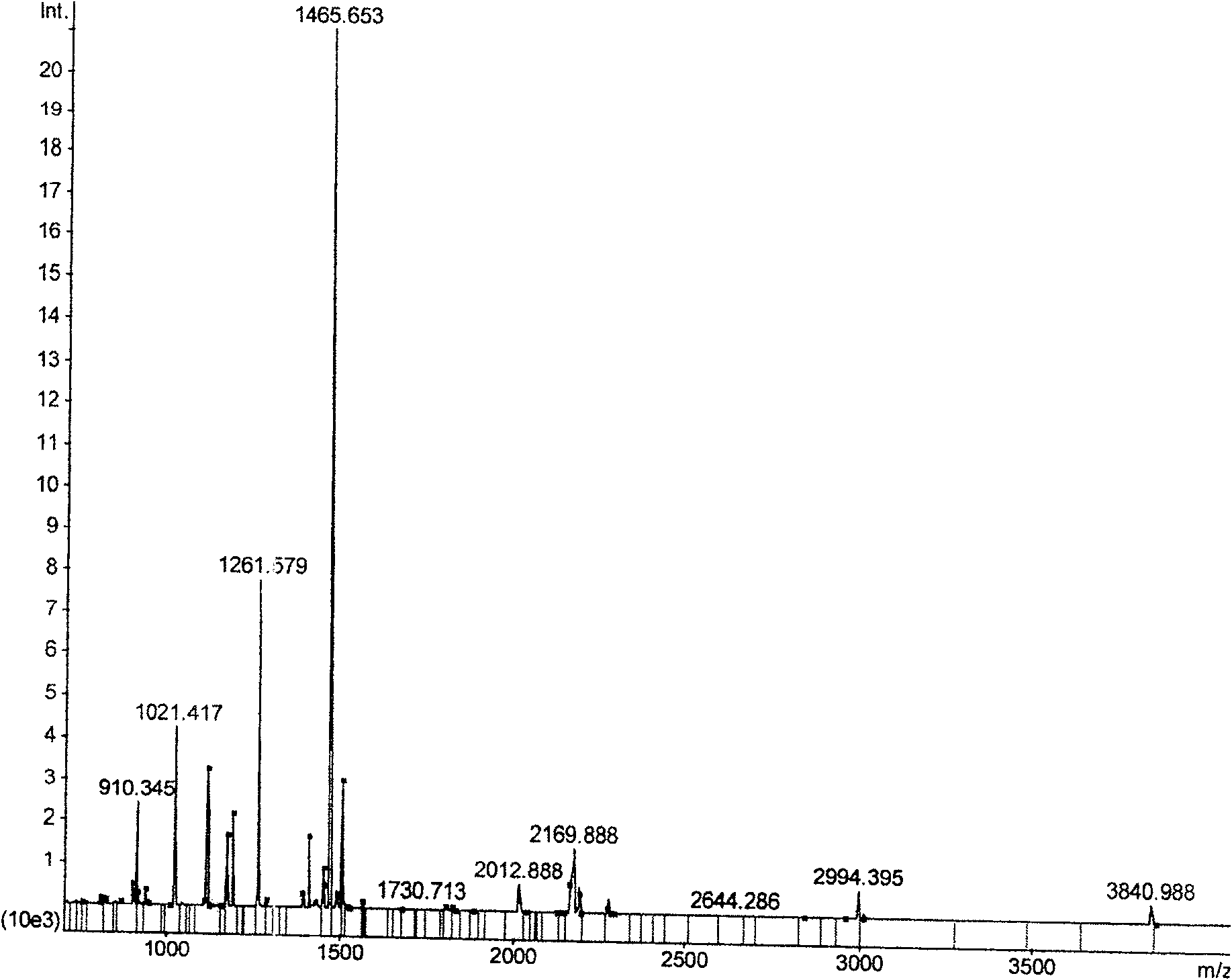
[0062] Example 2. Simplified in-gel enzymatic hydrolysis method for enzymatic hydrolysis of bovine serum albumin (BSA) and its effect verification
[0063] Bovine serum albumin (BSA) is a protein that is very abundant in bovine serum. In the NCBI database, there is a large amount of data information for reference, which greatly reduces the difficulty of identifying the protein by mass spectrometry. In order to quantitatively compare the K method and the simplified enzymatic hydrolysis method, a further comparative study was carried out with BSA as the experimental material.
[0064] Take 10μg-5ng BSA for one-dimensional electrophoresis separation, and different amounts of BSA are separated by one-dimensional SDS-PAGE ( Figure 2A ), the protein band with a content of 0.1 μg was cut manually ( Figure 2A , the band indicated by the arrow), and divided into 10 equal parts on average, then each part of gel contains 10ng bovine serum albumin (the weight of the gel containing this...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com