Method for extending DNA sequence by cyclical crosses
A DNA sequencing and DNA sequence technology, applied in the biological field, can solve the problems of increasing the information of erroneous extension, accumulating the amount of markers, and reducing the number of templates, so as to increase the information of erroneous extension, the determination of the sequence is accurate, and the cumulative effect is not serious. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
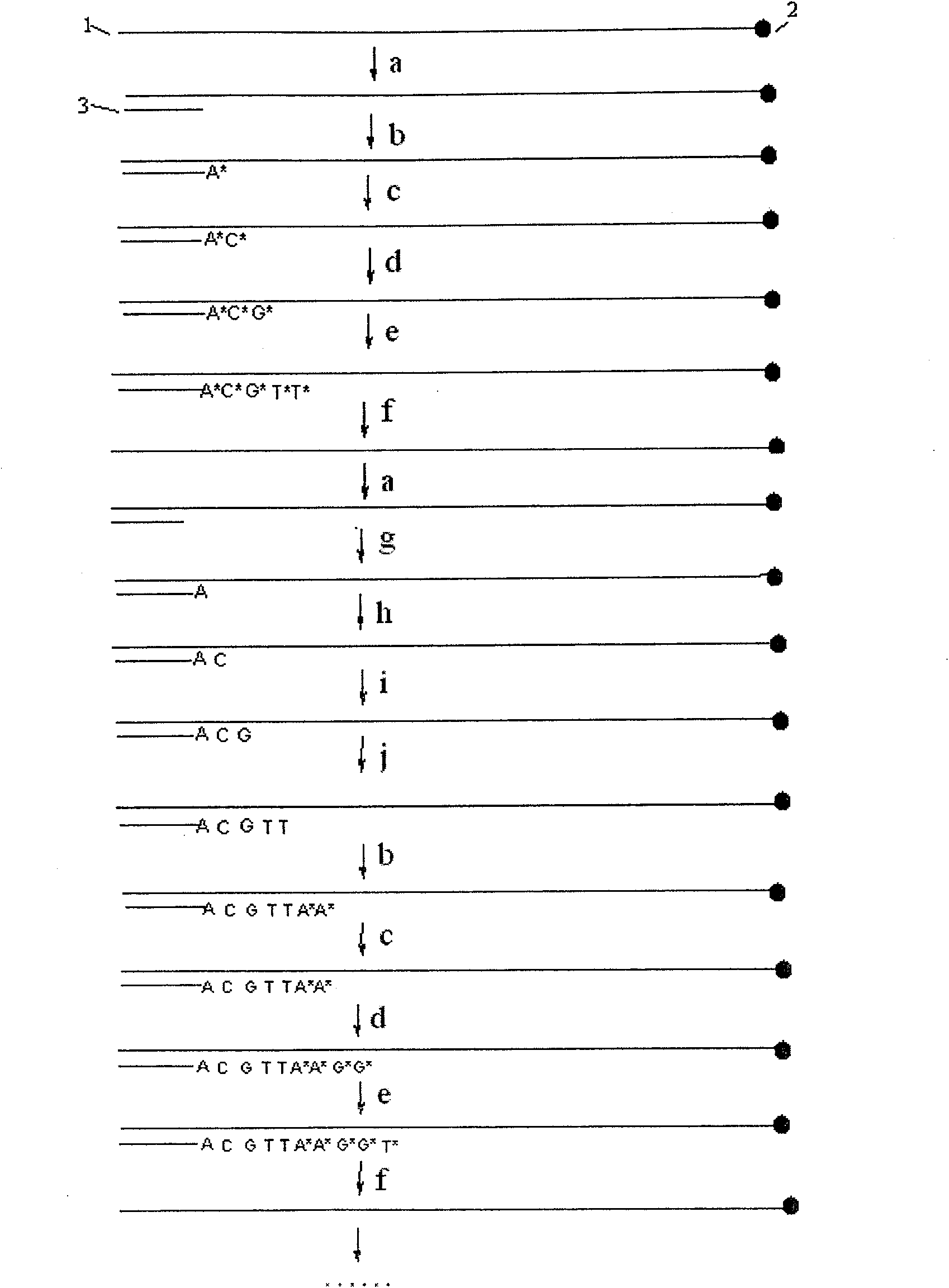
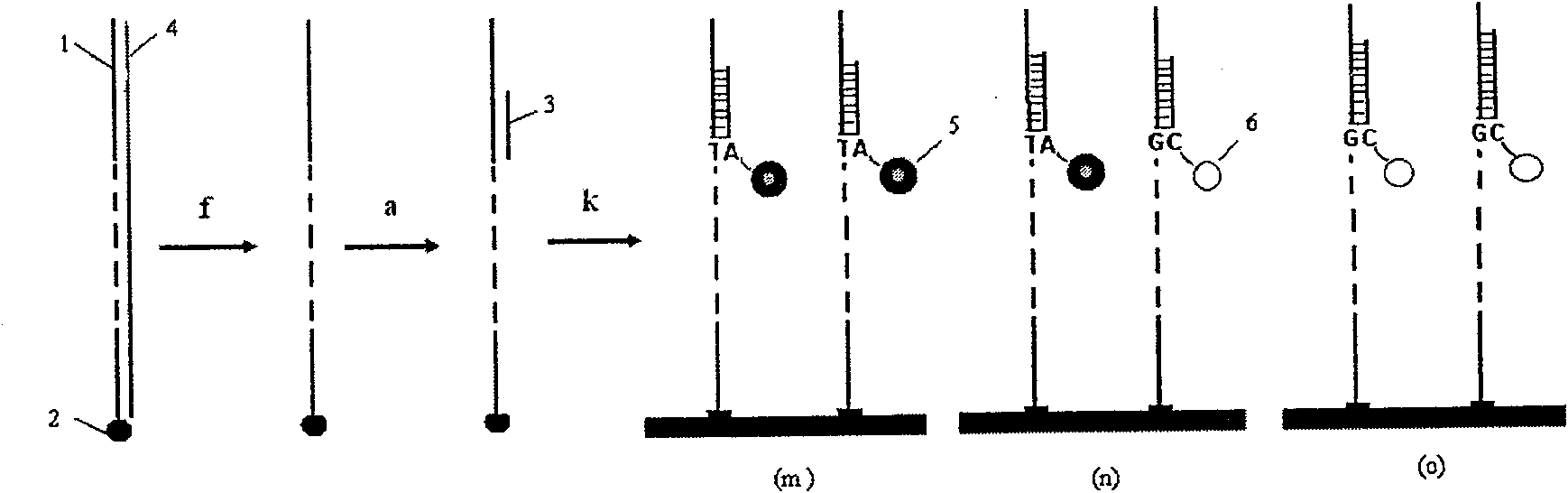
[0022] Example 1: The cycle hybridization-fluorescence extension sequencing method determines a DNA fragment containing the SNP site number rs11053646 in the human genome,
[0023] Refer to attached figure 1 , Design a pair of PCR primers, one of which is modified with acrylamide group. PCR primers are used to amplify a DNA sequence fragment containing SNP site number rs 11053646 in human samples (such as blood, saliva, etc.). The PCR product is fixed on a glass slide by polymerization method, and the unfixed PCR chain and other impurities are removed by denaturation and electrophoresis methods to obtain a pure DNA chain. One primer, unmodified, hybridizes to the immobilized DNA template.
[0024] Under the extension reaction conditions, according to the order of A, G, C, T, one base is added each time, and the fluorescently labeled monomer is added for one round, until the change in fluorescence is added each time after the labeled monomer is added and the polymerization is...
example 2
[0027] Example 2: Determination of a DNA fragment comprising the SNP site number rs11053646 in the human genome by cycle hybridization-pyrosequencing.
[0028] Refer to attached figure 1 , prepare the sequencing template according to the method of Example 1, and complete the hybridization with the sequencing primer.
[0029] According to the order of A, G, C, T, add one base each time and add unlabeled monomers respectively, and judge whether the extension and the light intensity of the base occur according to whether the light and light intensity are generated in the pyrosequencing system under the extension reaction conditions The number of extensions, so it can be inferred that the DNA template sequence determined in this hybridization is AAG ACT GGA TCTGGC ATG GAG AAA ACT G. When the amount of light change cannot reflect the number of extended bases, the pyrosequencing of this hybridization is stopped.
[0030] The sequencing primers extending the above 28 bases were den...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com