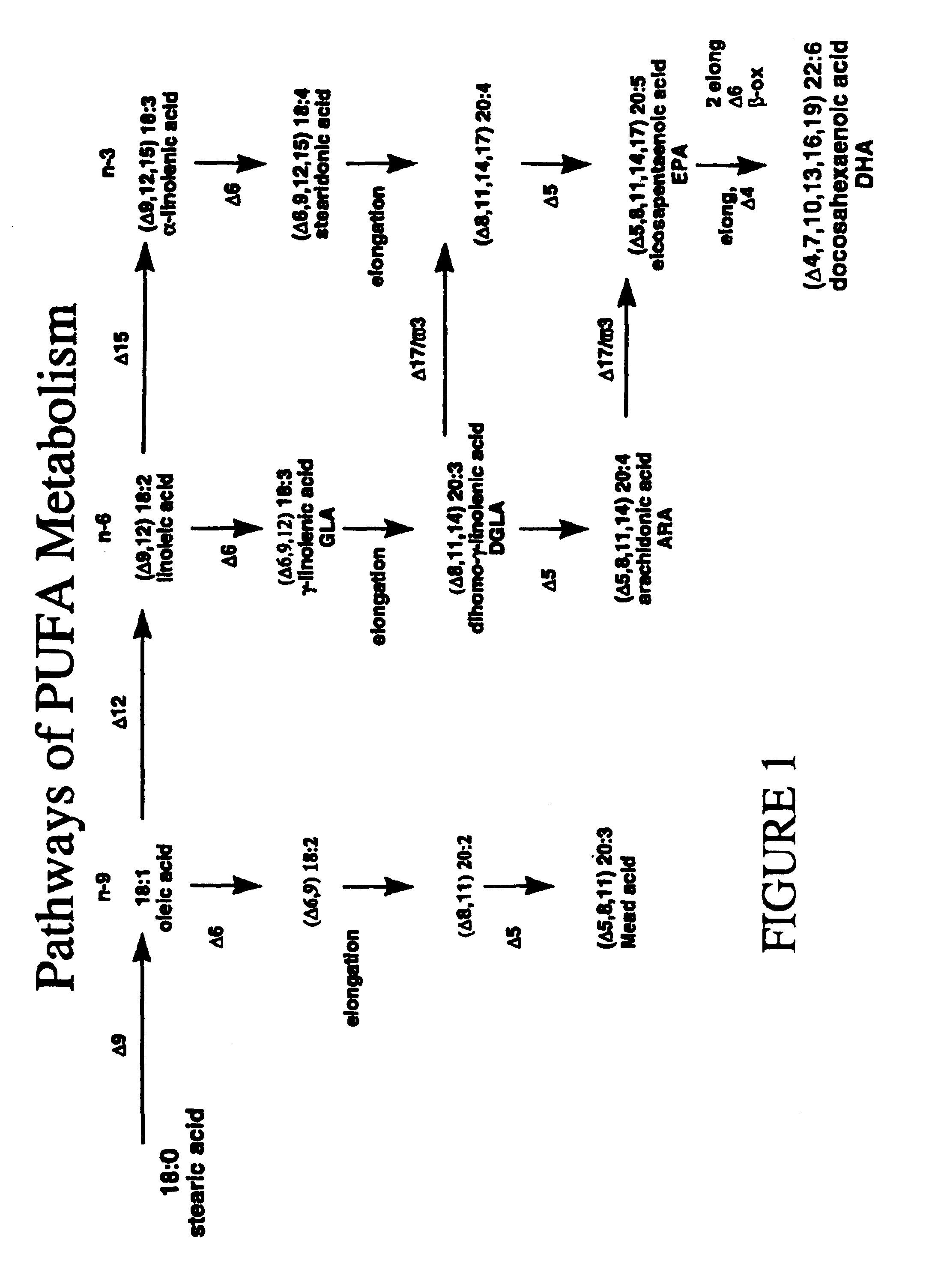
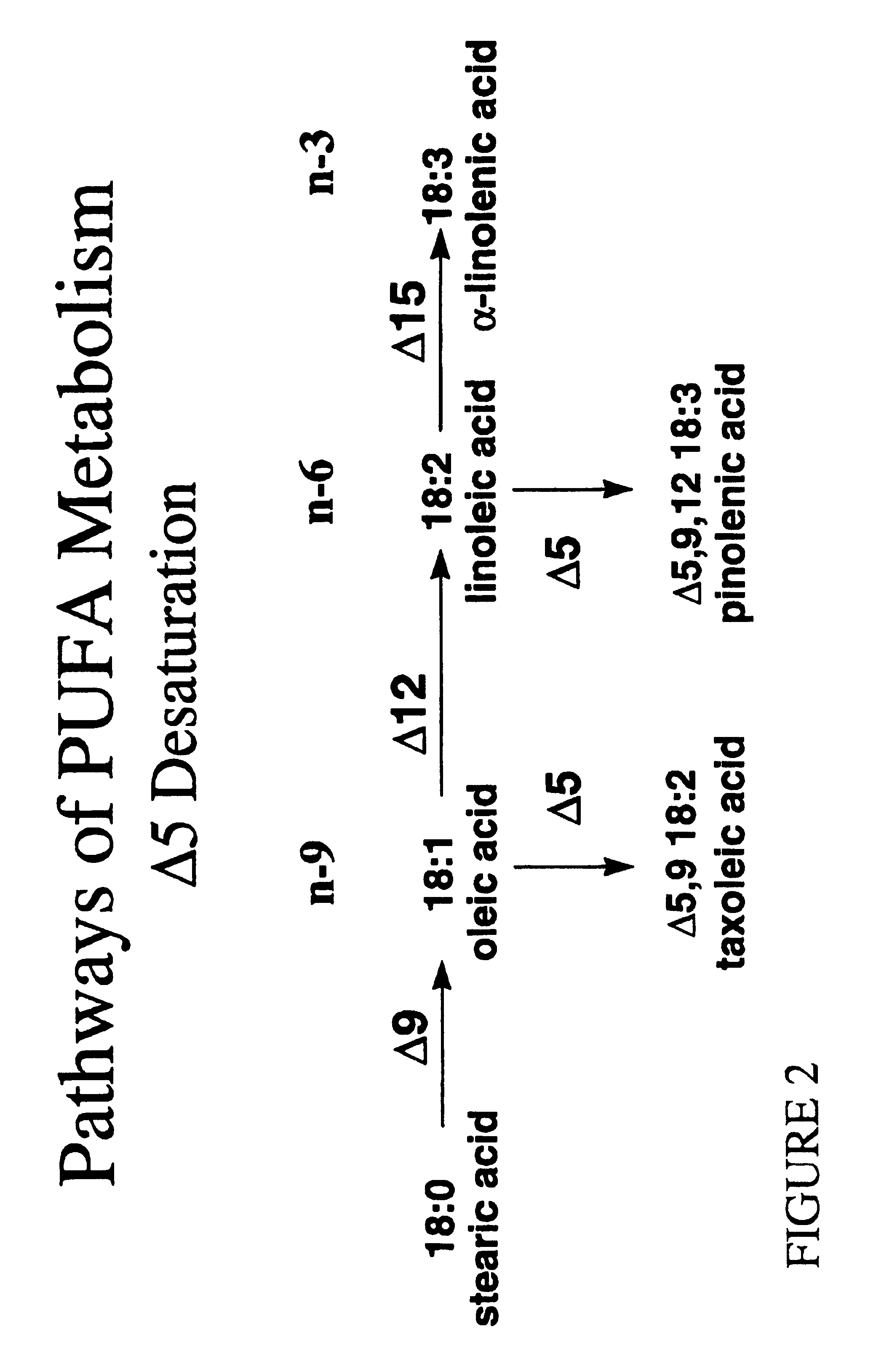
Polyunsaturated fatty acids in plants
a technology plants, applied in the field of polyunsaturated fatty acids in plants, can solve the problems of uncontrollable fluctuations in availability of natural sources, depletion of fish stocks, and natural variation or depletion of fish stocks, and achieve the effect of modifying the fatty acid profil
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Expression of ω-3 Desaturase from C. Elegans in Transgenic Plants
[0047]The Δ15 / ω-3 activity of Brassica napus can be increased by the expression of an ω-3 desaturase from C. elegans. The fat-1 cDNA clone (Genbank accession L41807; Spychalla, J. P., Kinney, A. J., and Browse, J. 1997 P.A.A.S. 94, 1142-1147, SEQ ID NO:1 and SEQ ID NO:2) was obtained from john Browse at Washington State University. The fat-1 cDNA was modified by PCR to introduce cloning sites using the following primers:[0048]Fat-1forward (SEQ ID NO:3):[0049]5′-CUACUACUACUACTGCAGACAATGGTCGCTCATTCCTCAGA-3′[0050]Fat-1reverse (SEQ ID NO:4):[0051]5′-CAUCAUCAUCAUGCGGCCGCTTACTTGGCCTTTGCCTT-3′
[0052]These primers allowed the amplification of the entire coding region and added PstI and NotI sites to the 5′- and 3′-ends, respectively. The PCR product was subcloned into pAMP1 (GIBCOBRL) using the CloneAmp system (GIBCOBRL) to create pCGN5562. The sequence was verified by sequencing of both strands to be sure no changes were intro...
example 2
Over-Expression of Δ15-desaturase Activity in Transgenic Canola
[0060]The Δ15-desaturase activity of Brassica napus can be increased by over-expression of the Δ15-desaturase cDNA clone.
[0061]A. B. napus Δ15-desaturase cDNA clone was obtained by PCR amplification of first-strand cDNA derived from B. napus cv. 212 / 86. The primers were based on published sequence: Genbank #L01418 Arondel et al, 1992 Science 258:1353-1355 (SEQ ID NO:7 and SEQ ID NO:8).
The following primers were used:
[0062]Bnd15-FORWARD (SEQ ID NO:9)[0063]5′-CUACUACUACUAGAGCTCAGCGATGGTTGTTGCTATGGAC-3′[0064]Bnd15-REVERSE (SEQ ID NO:10)[0065]5′-CAUCAUCAUCAUGAATTCTTAATTGATTTTAGATTTG-3′
[0066]These primers allowed the amplification of the entire coding region and added SacI and EcoRI sites to the 5′- and 3′-ends, respectively The PCR product was subcloned into pAMP1 (GIBCOBRL) using the CloneAmp system (GIBCOBRL) to create pCGN5520. The sequence was verified by sequencing of both strands to be sure that the open reading frame ...
example 3
Expression of Δ5 Desaturase in Plants Expression in Leaves
[0072]Ma29 is a putative M. alpina Δ5 desaturase as determined by sequence homology (SEQ ID NO:11 and SEQ ID NO:12). This experiment was designed to determine whether leaves expressing Ma29 (as determined by Northern) were able to convert exogenously applied DGLA (20:3) to ARA (20:4).
[0073]The Ma29 desaturase cDNA was modified by PCR to introduce convenient restriction sites for cloning. The desaturase coding region has been inserted into a d35 cassette under the control of the double 35S promoter for expression in Brassica leaves (pCGN5525) following standard protocols (see U.S. Pat. No. 5,424,200 and U.S. Pat. No. 5,106,739). Transgenic Brassica plants containing pCGN5525 were generated following standard protocols (see U.S. Pat. No. 5,188,958 and U.S. Pat. No. 5,463,174).
[0074]In the first experiment, three plants were used: a control, LPO04-1, and two transgenics, 5525-23 and 5525-29. LP004 is a low-linolenic Brassica var...
PUM
| Property | Measurement | Unit |
|---|---|---|
| weight percent | aaaaa | aaaaa |
| weight percent | aaaaa | aaaaa |
| weight percent | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com