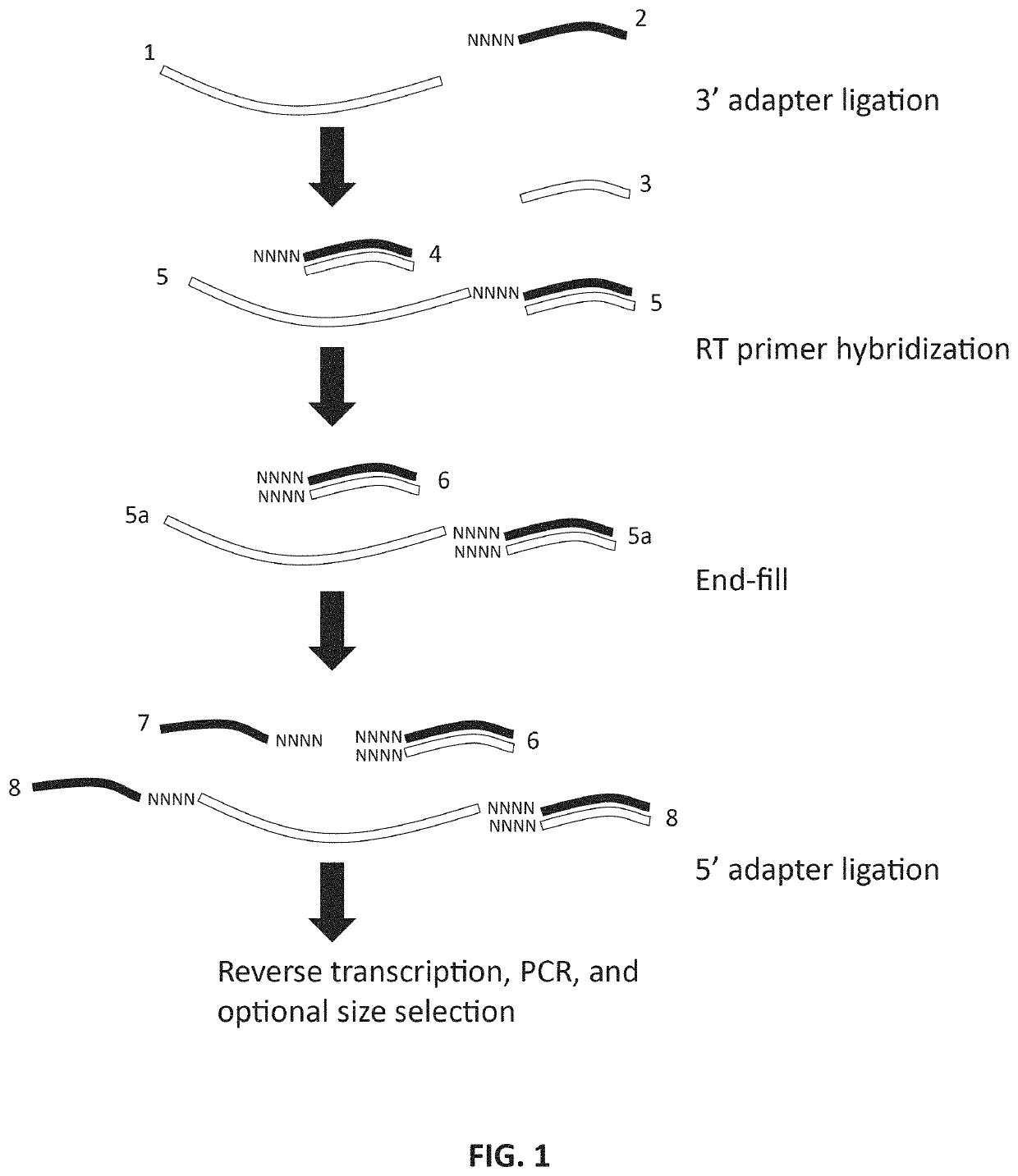
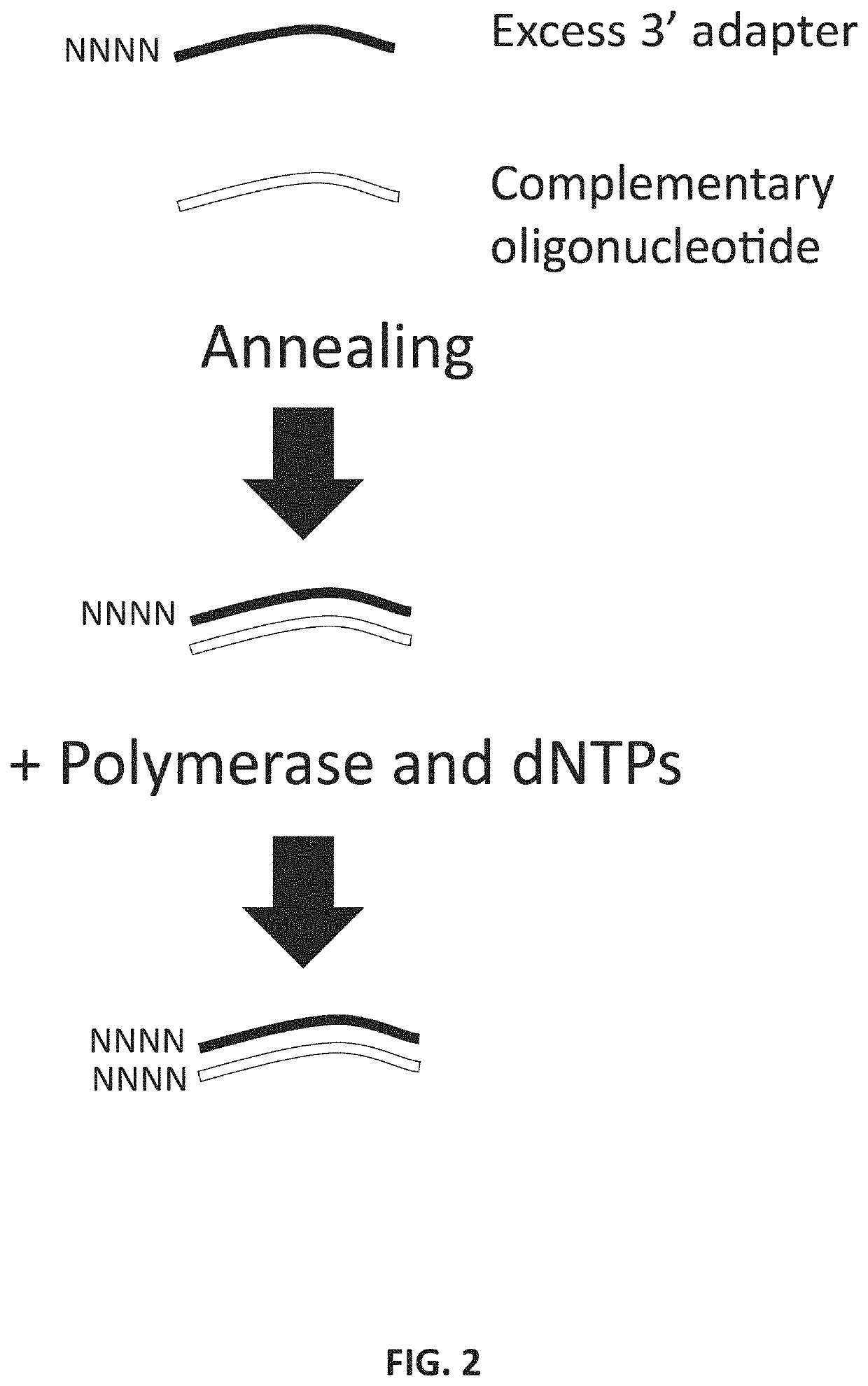
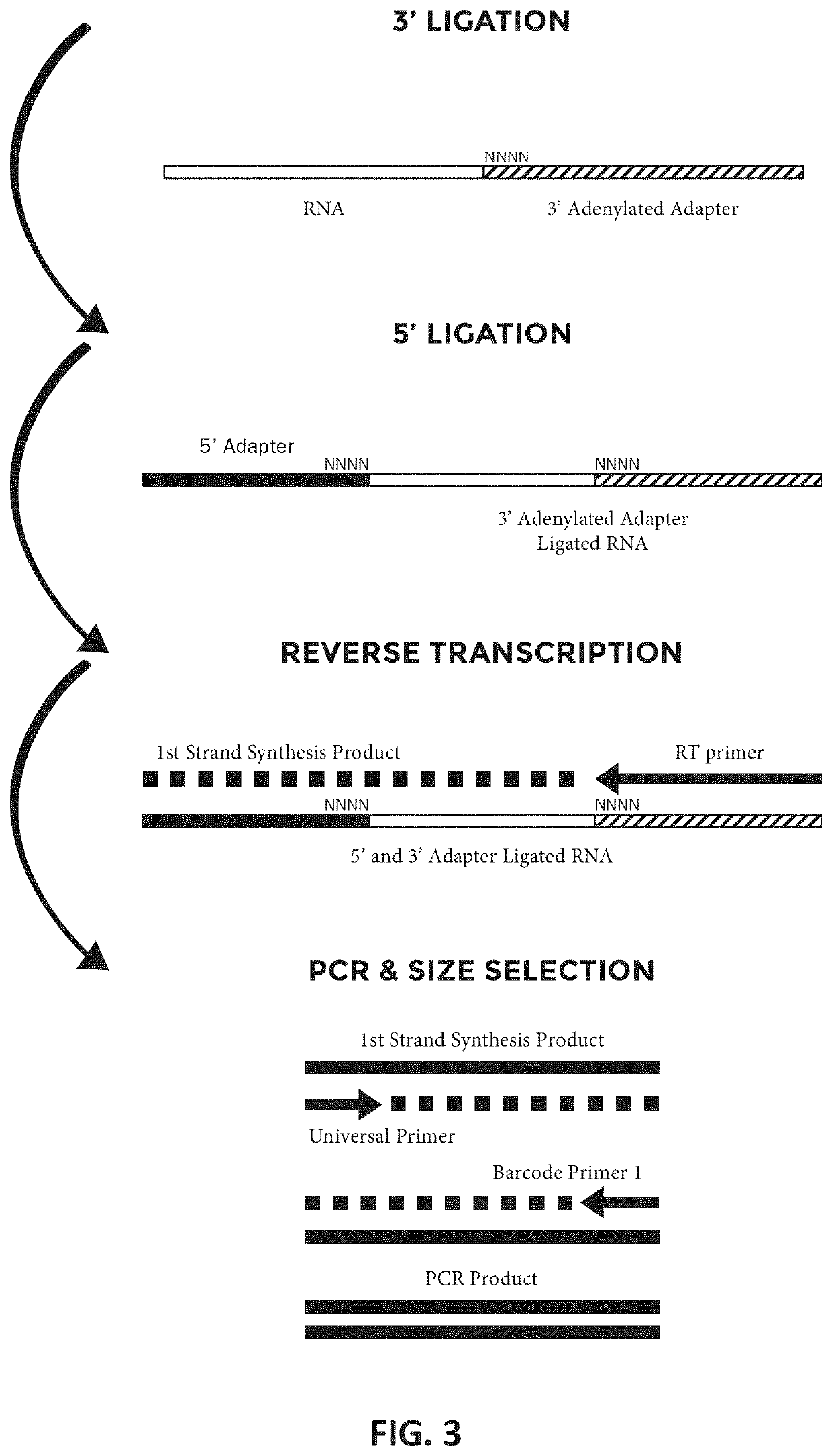
Methods and kits for reducing adapter-dimer formation
a technology of adapter dimer and kit, which is applied in the field of reducing adapter dimer formation, can solve the problems of severe bias, inability to select spri size, and inability to gel-based purification,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of an Exemplary SRNA-Seq Library
[0053]In an exemplary method embodiment, sRNA-seq libraries were prepared from human brain total RNA (Ambion, cat. # AM7962) using the NEXTFLEX™ Small RNA Sequencing Kit v2 according to manufacturer's instructions, except as indicated. For sRNA-seq libraries prepared according to certain disclosed methods, following Excess 3′ Adapter Removal using NEXTFLEX™ beads, 11.5 μL supernatant of nuclease free water containing 3′ ligation products was recovered. To each supernatant 1.5 μL of NEBuffer 2.1 (500mM NaCl, 100 mM Tris-HCl, 100 mM MgCl2, 1 ng / ml BSA, pH 7.9), 0.5 uL of 6.25 uM dNTPs, and 0.5 uL of T4 DNA Polymerase (Enzymatics, Cat. # P7080L) were added and incubated at 12° C. for 15 minutes followed by 50 ° C. for 20 minutes. The following modifications were then made to the NEXTFLEX™ Small RNA sequencing kit v2 protocol: 1) in the 5′ adapter ligation step, samples were not heated at 70° C. for 2 minutes. Instead, the 5′ 4N adapter was h...
example 2
Effectiveness of End-Filling in Reducing Adapter-Dimers
[0054]To show the effectiveness of the disclosed end-filling method, sRNA-seq libraries were prepared from 100 ng human brain total RNA, as described in Example 1, either with or without the described end-filling method and analyzed by TBE-PAGE (FIG. 5). Lanes 1 and 2 are technical duplicates of small RNA libraries created with the proposed end-fill method and lanes 3 and 4 are technical duplicates of small RNA libraries created without the end-fill method; lane M contains a base pair ladder standard. The results demonstrate that the method is not only effective in reducing adapter-dimer but surprisingly also increases yield of insert-containing product.
example 3
Effectiveness of Annealing RT-Primer Prior to 3′ Ligation
[0055]3′ adapter was pre-annealed to oligonucleotide and libraries were prepared from 100 ng human brain total RNA, as described in Example 1, using either the pre-annealed oligonucleotide-3′ adapter duplexes or with the oligonucleotide annealed after 3′ ligation. Referring to FIG. 6, lanes 1-4 depict small RNA libraries prepared with annealing of the oligonucleotide to the 3′ adapter prior to the 3′ ligation step (“Pre-anneal”); and lanes 5-8 depict small RNA libraries prepared by annealing the oligonucleotide to the 3′ adapter after to the 3′ ligation step (“Post-anneal”). Lanes 1,2,5, and 6 depict libraries prepared according to the current teachings; while the libraries depicted in lanes 3,4,7, and 8 depict libraries were prepared without the end-filling technique of the current teachings; lanes marked M contain a base pair ladder standard. The results demonstrate that pre-annealing the 3′ adapter and the oligonucleotide d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com