Method for the quantification of pd-l1 expression
a pdl1 and expression technology, applied in the field of pdl1 expression quantification, can solve the problems of t-cell exhaustion, ineffective response, and many patients who do not benefit from these therapies, and achieve the effect of reducing variations and increasing errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Development and Analytical Validation of the Assay
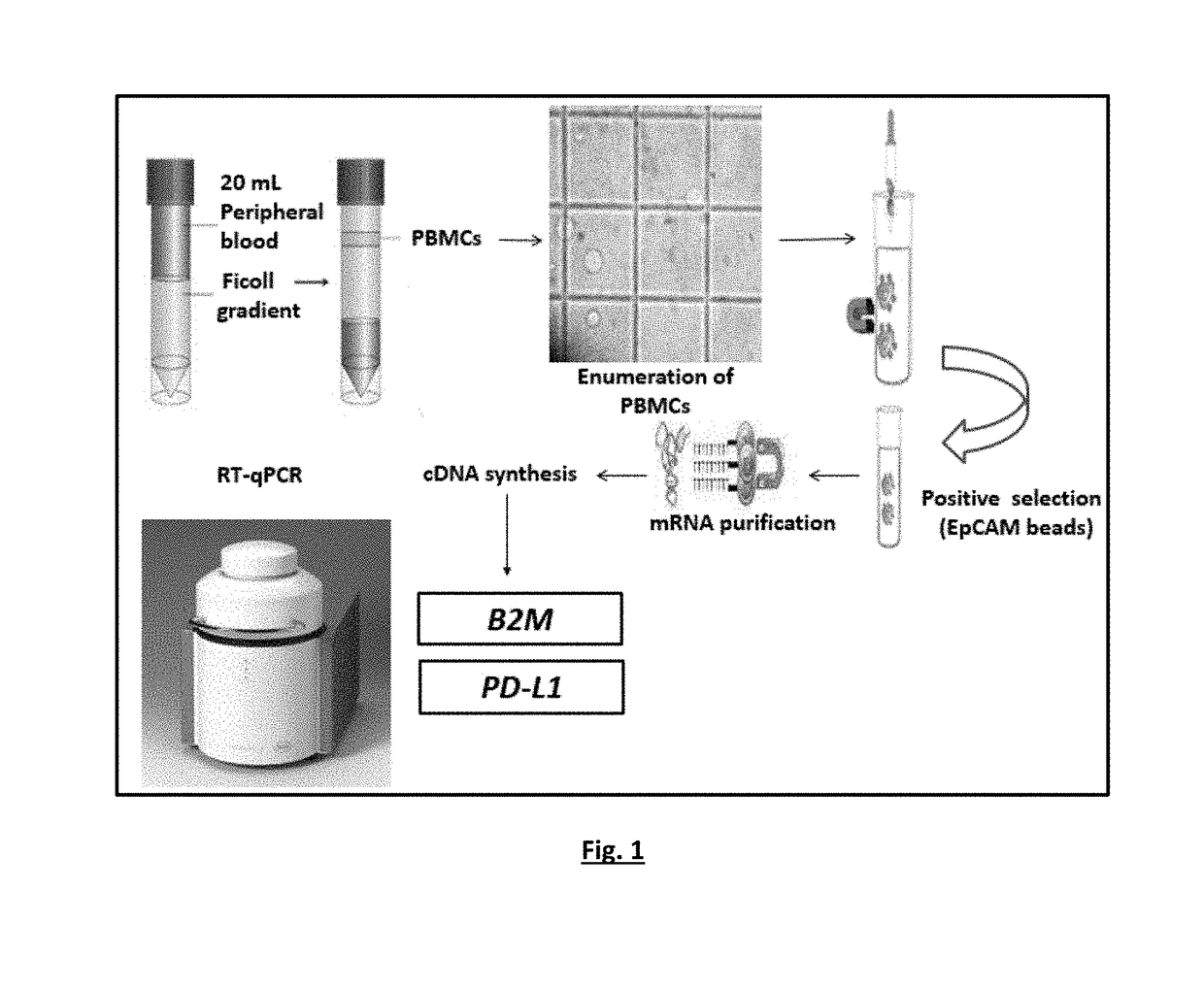
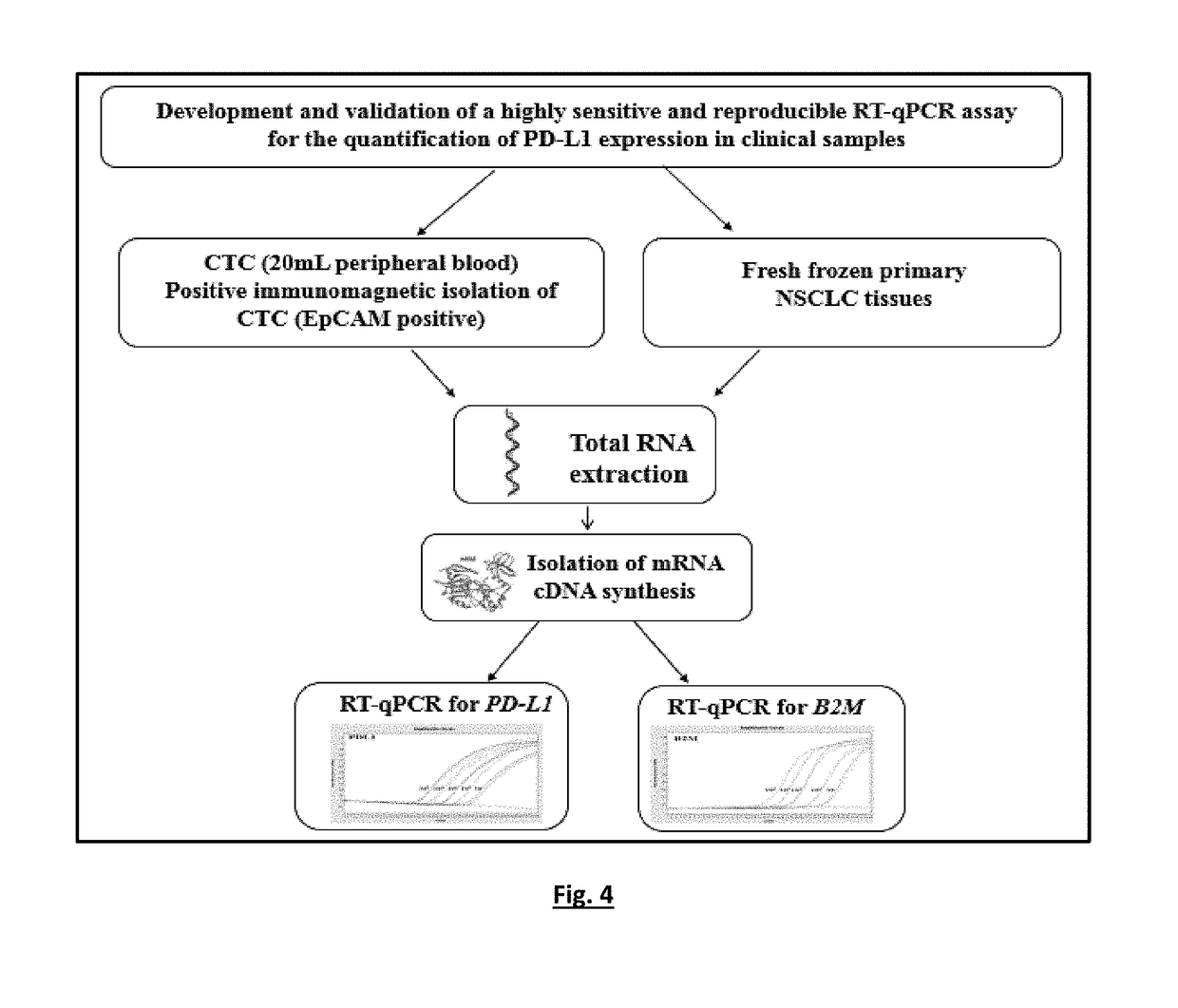
[0135]The experimental flowchart of the study is outlined in FIG. 4.
Protocol Optimization.
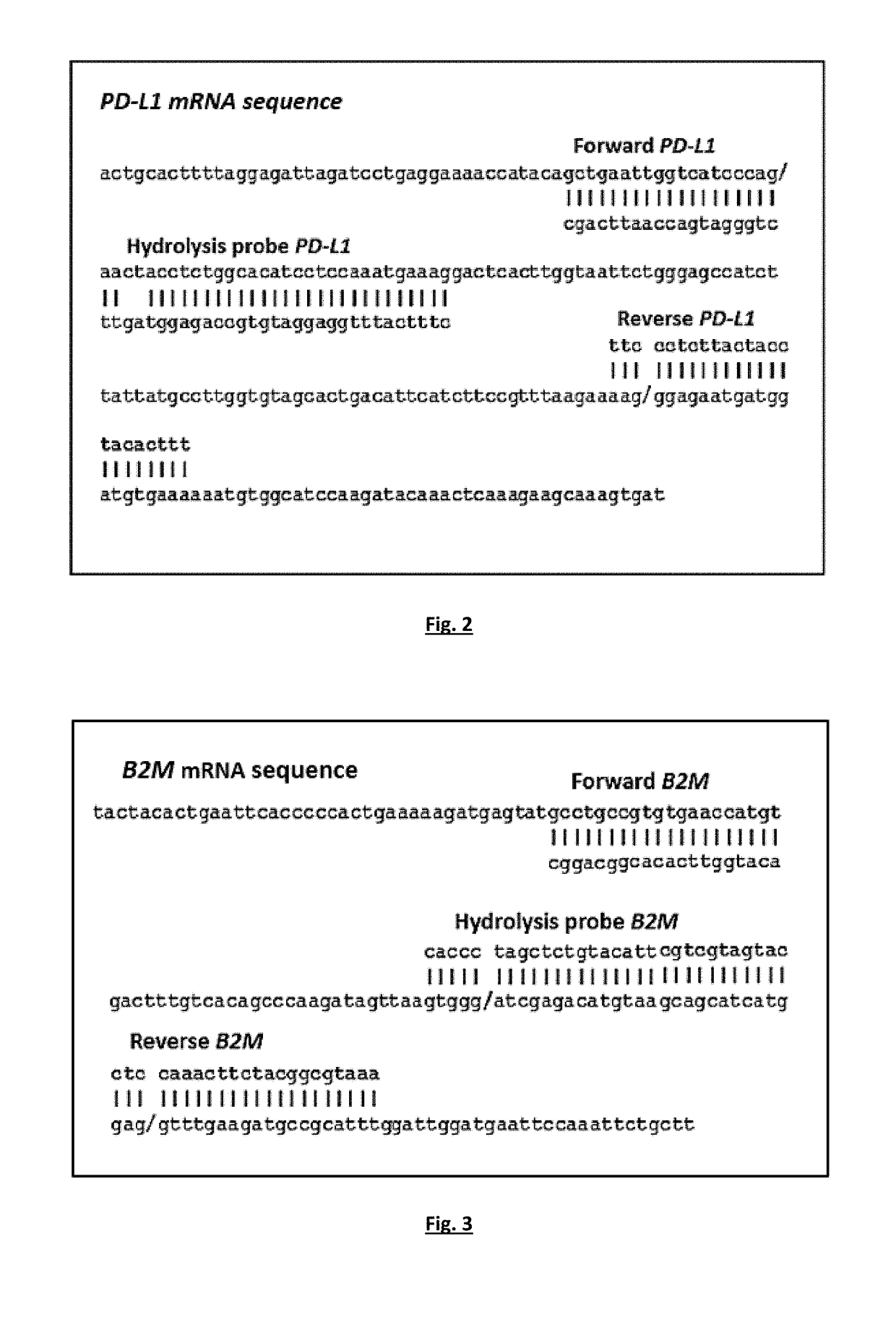
[0136]A RT-qPCR based methods for the quantification of PD-L1 and B2M expression were optimized in a number of experiments, using as positive control cDNA samples from peripheral blood mononuclear cells (PBMC) of healthy control samples and negative control of PCR reaction mix, with respect to: PCR annealing temperature, cycling parameters, primers and probe concentrations, Mg+2 and dNTPs concentrations.
[0137]Single RT-qPCR was performed for PD-L1 and B2M expression. Quantification is based on real-time monitoring during PCR of labelled specific hydrolysis probe for PD-L1. The cycle where the fluorescence signal rises above background noise (quantification cycle, Cq) is best quantified through the LightCycler software as the second derivative maximum of the curve. Real-time RT-PCR for PD-L1 mRNA was performed using the LightCycler system (Roche ...
example 2
RT-qPCR Quantification Using External Calibrators
[0140]Firstly, individual PCR amplicons specifically for PD-L1 and B2M were generated in order to be used as external quantification calibrators. For this purpose, total RNA was extracted from PBMC pool from normal samples since PBMC express as well PD-L1. cDNA was synthesized and served as a template for the amplification of PD-L1 and B2M by the above described RT-qPCR. PCR products were purified using MinElute PCR Purification Kit (Qiagen, Germany) and the amplicons were quantified in the Nanodrop-1000 spectrophotometer (NanoDrop, Technologies, USA).
[0141]DNA concentration was converted to copies / μL by use of the Avogadro number and the molecular weight of the amplicon number of bases of the PCR product multiplied by the mean molecular weight of a pair of nucleic acids which is 660. A standard stock solution corresponding to 1010 copies / μL for each gene transcript was prepared. Serial dilutions of this stock amplicon solution in DNa...
example 3
The Limit of Detection and Linearity of Assay
[0142]The limit of detection (LOD) of the developed RT-qPCR assay for both PD-L1 and B2M as copies / μL in the reaction was evaluated. To estimate LOD, the quantification calibrators containing a known number of copies / μL were prepared as described below in detail: For each gene target a calibration curve was generated using serial dilutions of these standards in triplicate for each concentration, ranging from 105 copies / μL to 10 copies / μL. The calibration curves showed linearity from 10 copies / μL up to 105 copies / μL, with correlation coefficients larger than 0.99 in both cases, indicating a precise log-linear relationship (FIG. 5). None of the primers and hydrolysis probes gave any signal for any of the gene target transcripts when 50 ng / μL of 10 different genomic DNAs was analyzed.
[0143]The LOD for both of these assays was found to be 3 copies / μL and the limit of quantification (LOQ) was found to be 9 copies / μL, (estimated according to th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com