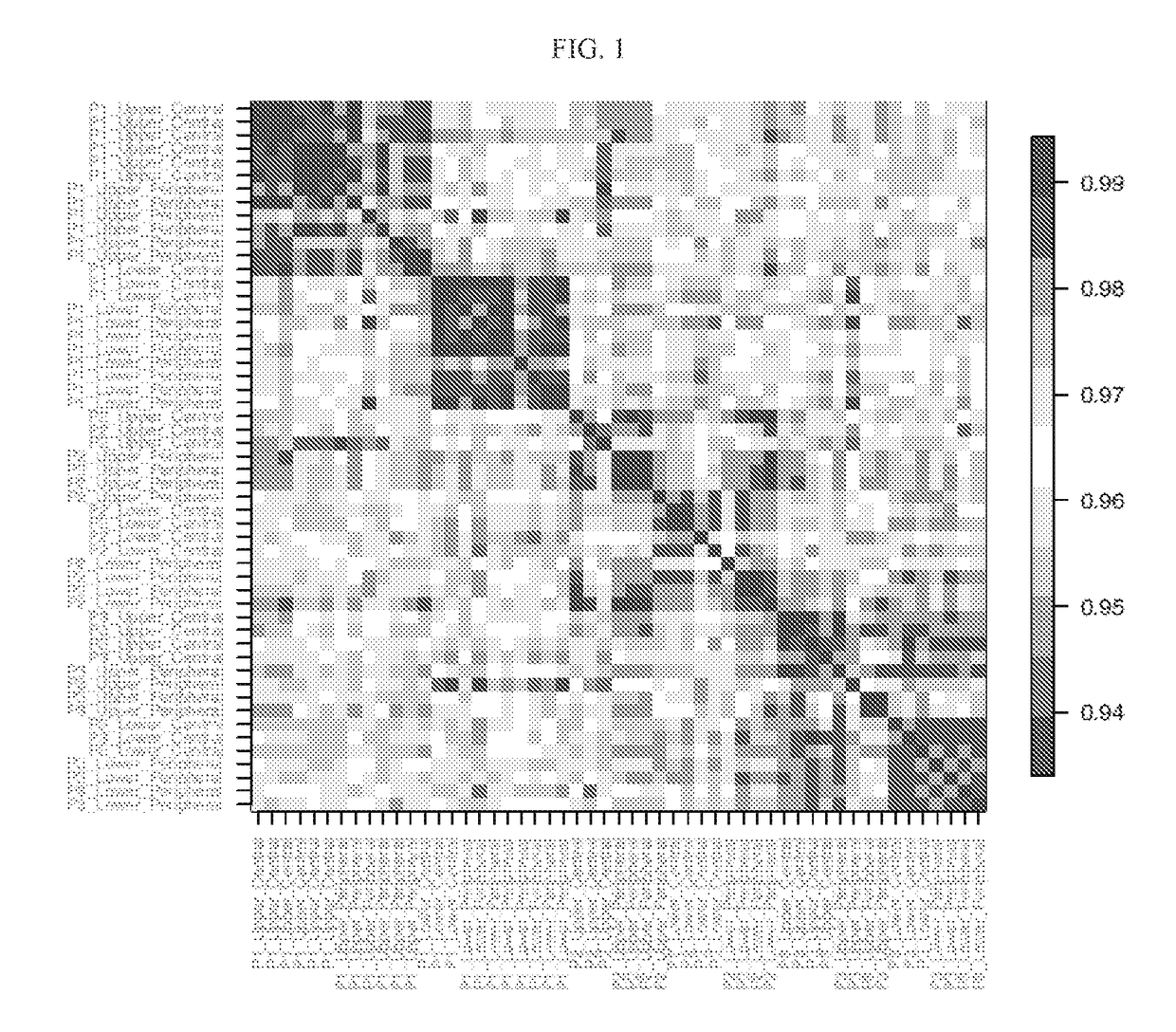
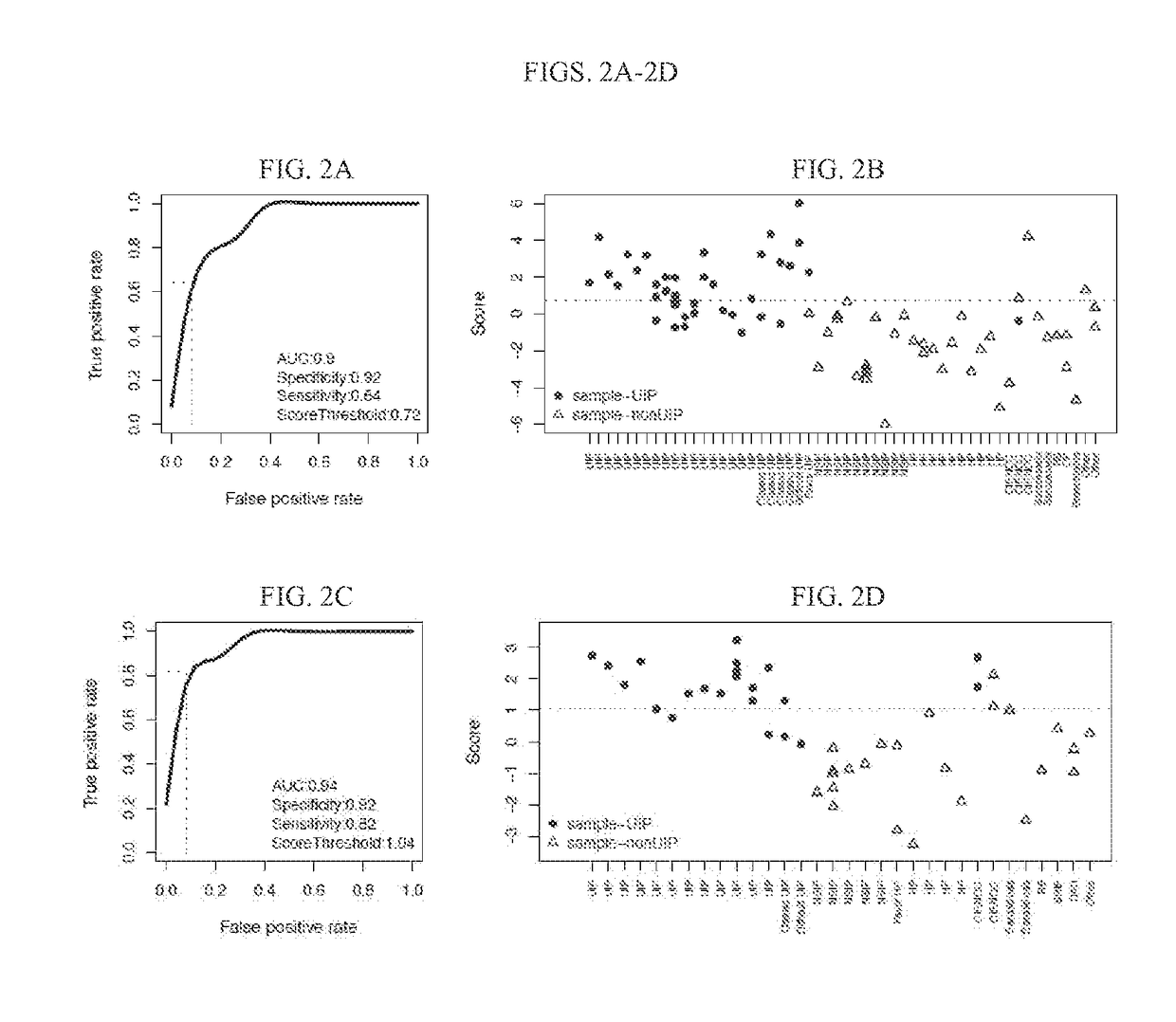
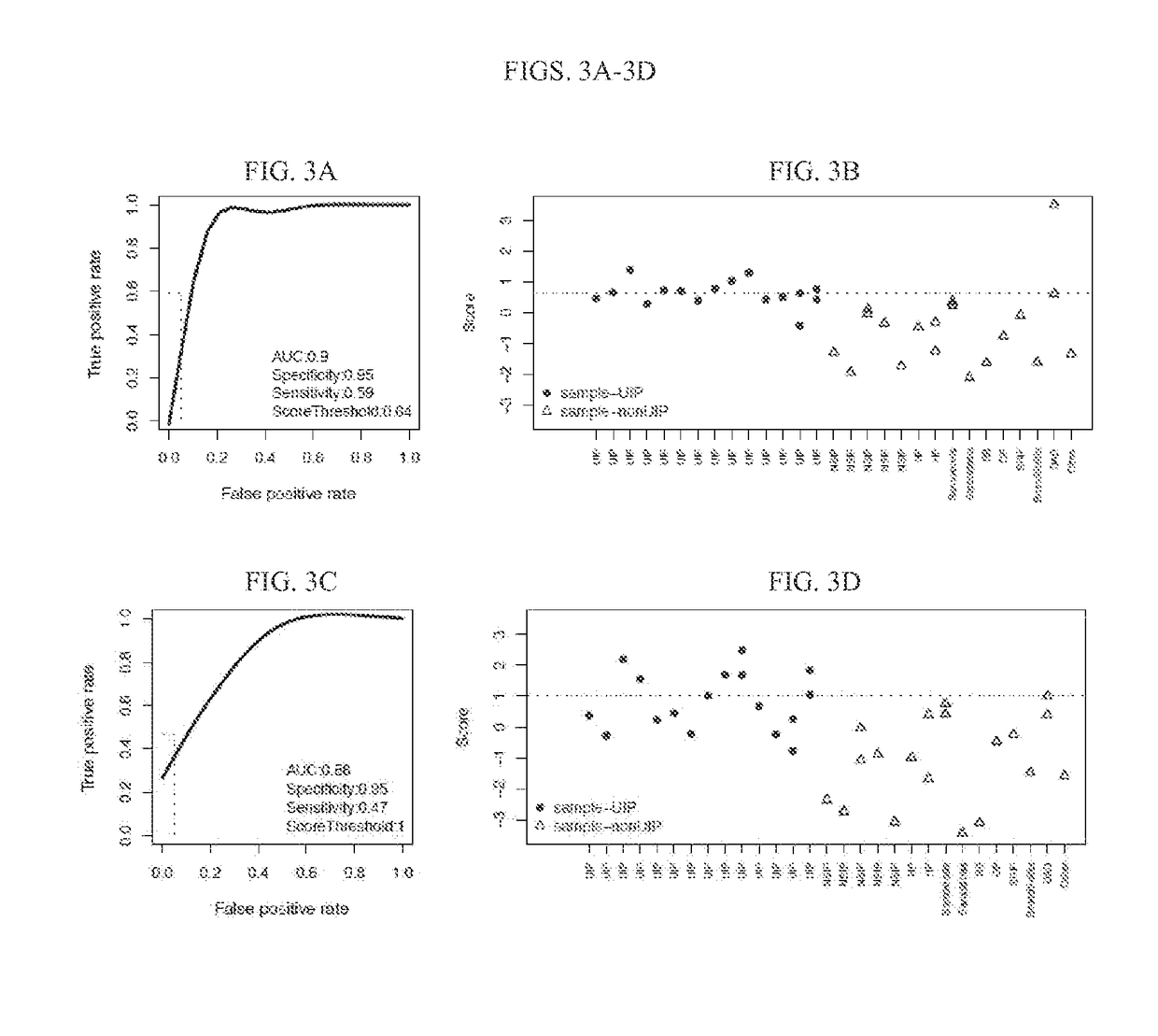
Systems and methods of diagnosing idiopathic pulmonary fibrosis on transbronchial biopsies using machine learning and high dimensional transcriptional data
a technology of pulmonary fibrosis and biopsies, applied in the field of systems and methods of diagnosing idiopathic pulmonary fibrosis on transbronchial biopsies using machine learning and high dimensional transcriptional data, can solve the problems of difficult diagnosis of ipf, time-consuming review, and inability to definitively diagnose ip
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sample Collection, Pathology Diagnosis, and Labeling
[0233]Video-assisted thoracoscopic surgery (VATS) specimens were prospectively collected as a part of an Institutional Review Board (IRB) approved ongoing multi-center clinical protocol, BRonchial sAmple collection for a noVel gEnomic test (BRAVE), sponsored by Veracyte, Inc. (South San Francisco, Calif.). Additional VATS and surgical lung biopsy specimens were obtained from banked sources.
[0234]Following surgery, histology slides were collected, de-identified, and submitted to expert pathology review. Selected slides were scanned to construct a permanent digital file of microscopic images (Aperio, Vista, Calif.). Slides were evaluated according to the central pathology diagnostic process described in FIG. 5, resulting in both sample-level and patient-level pathology diagnoses. Pathology categories are summarized in Table 3. A patient can have more than one sample-level diagnosis (i.e. one per VATS sample per patient, most often on...
example 2
[0237]Frozen tissue samples were mounted for sectioning using Tissue-Tek O.C.T. medium (Sakura Finetek U.S.A.) and 2×20 μm sections generated using a CM1800 cryostat (Leica Biosystems, Buffalo Grove, Ill.). Tissue curls were immediately immersed in RNAprotect (QIAGEN, Valencia, Calif.), incubated overnight at 4° C. and stored at −80° C. until extraction. Whenever possible, adjacent 5 μm tissue curls were mounted onto glass slides and processed for hematoxylin and eosin (H&E) staining following standard procedures.
[0238]Nucleic acids were extracted using the AllPrep Micro Kit (QIAGEN) according to manufacturer's guidelines. Total RNA yield and quality was determined using Quant-it (Invitrogen) and Pico BioAnalyzer kits (Agilent). Fifteen nanograms of total RNA were amplified using Ovation FFPE WTA System (NuGEN, San Carlos, Calif.), hybridized to GeneChip Gene ST 1.0 (Affymetrix, Santa Clara, Calif.) microarrays, processed and scanned according to the manufacturer's ...
example 3
Next-Generation RNA Sequencing
[0239]Whole transcriptome RNA sequencing was performed on select samples at a targeted minimum read depth of 80 million paired-end reads per sample. Briefly, 10 ng of total RNA was amplified using the Ovation RNASeq System v2 (NuGEN, San Carlos, Calif.) and TruSeq (Illumina, San Diego, Calif.) sequencing libraries were prepared and sequenced on an Illumina HiSeq according to manufacturer's instructions. Raw reads were aligned to the hg19 genome assembly using TopHat2. Gene counts were obtained using HTSeq and normalized in Bioconductor using the varianceStabilizingTransformation function in the DESeq2 package. Raw counts and normalized expression levels were obtained for 55,097 transcripts.
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid amplification | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com