Modified human u1snrna molecule, a gene encoding for the modified human u1snrna molecule, an expression vector including the gene, and the use thereof in gene therapy of familial dysautonomia and spinal muscular atrophy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
n of the Modified U1 snRNAs
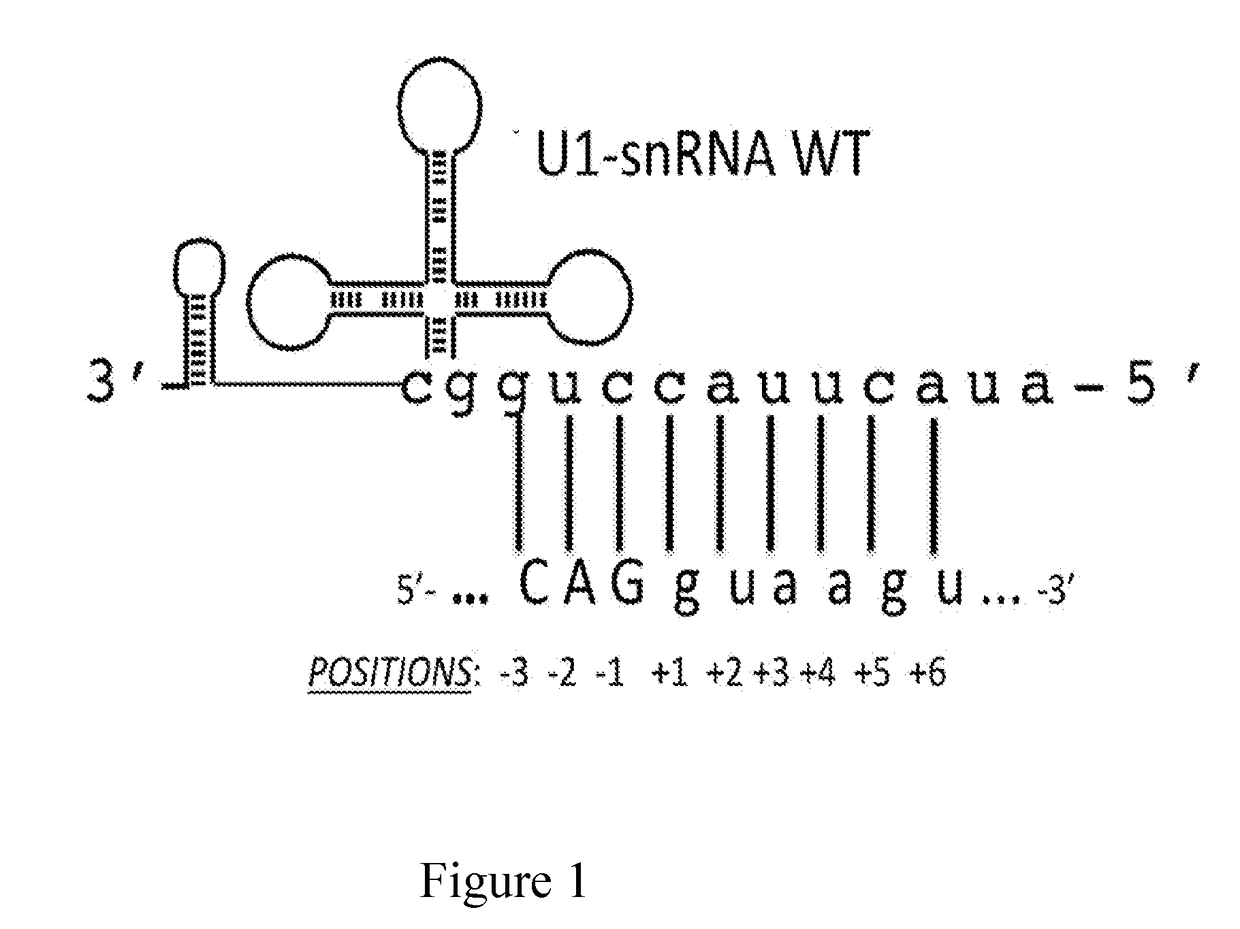
[0064]The modified U1 snRNAs were generated by the following procedure: the plasmid containing the sequence of the wild-type U1-snRNA gene, that is the non-modified U1-snRNA, was digested with the BglII and Bcll restriction enzymes. The sequence comprised between these two restriction sites was replaced with a double-stranded oligonucleotide comprising the binding sequence. The direct and reverse sequences of each oligonucleotide are described in Table 1 below and the resulting modified U1-snRNAs are named after the employed oligonucleotides.
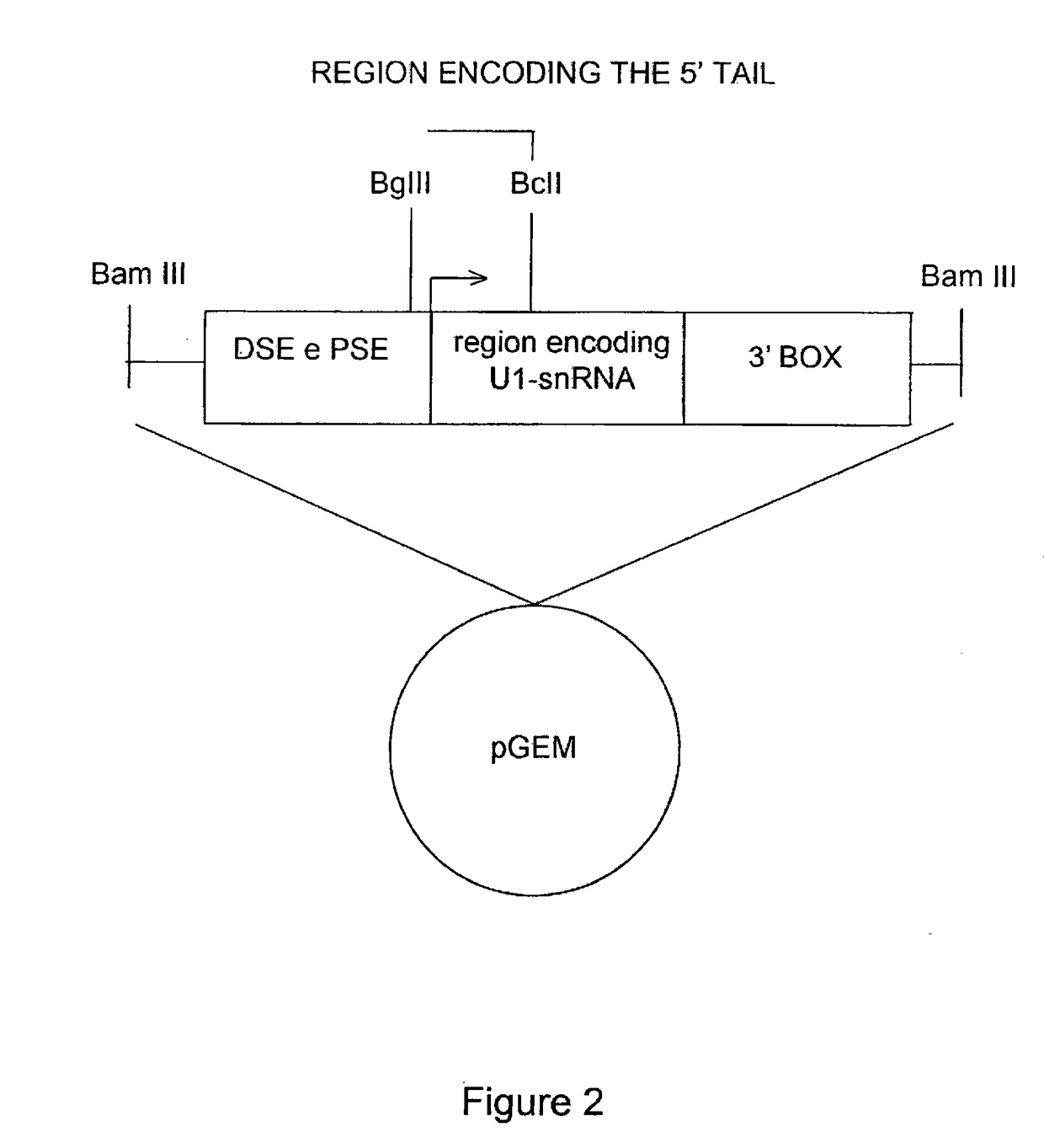
[0065]Furthermore, FIG. 2 shows a schematic representation of the U1 snRNA gene elements. The cloning strategy by which the different modified U1 snRNAs were prepared is indicated. FIG. 2 shows the U1snRNA gene with the promoter elements DSE and PSE, the region encoding for U1 snRNA (in the middle), and the 3′ processing box, inserted in a plasmid vector (pGEM). The transcription start site is indicated by an arrow. Th...
example 2
ion of the Minigenes into Cultured Cells and Analysis of the Splicing Products
[0066]The containing-vectors were inserted into the cells by transient transfection with Lipofectamine (liposomes). Following extraction of total cellular RNA with Trizol, the RNA was analyzed by RT-PCR with specific primers.
[0067]The reaction occurs in two steps: the RNA inverse transcription into a cDNA strand by a reverse transcriptase using random primers as templates, and amplification of the obtained cDNA by a DNA polymerase.
[0068]The PCR reaction was carried out in a final volume of 25 μl of a mixture containing:[0069]5 μl of AMV / Tfl 5× buffer suitable for the correct functioning of both the enzymes mentioned above;[0070]1 μl of 10 mM dNTPs mix;[0071]50 pmol of forward primer and 50 pmol of reverse primer;[0072]2 μl 25 mM MgSO4;[0073]2 μl of cell-extracted RNA;[0074]1 μl of AMV-RT (0.1 μ / μl), 1 μl of Tfl DNA polymerase;[0075]ultra pure H2O q.s.
[0076]The reverse transcription step was performed at 45...
example 3
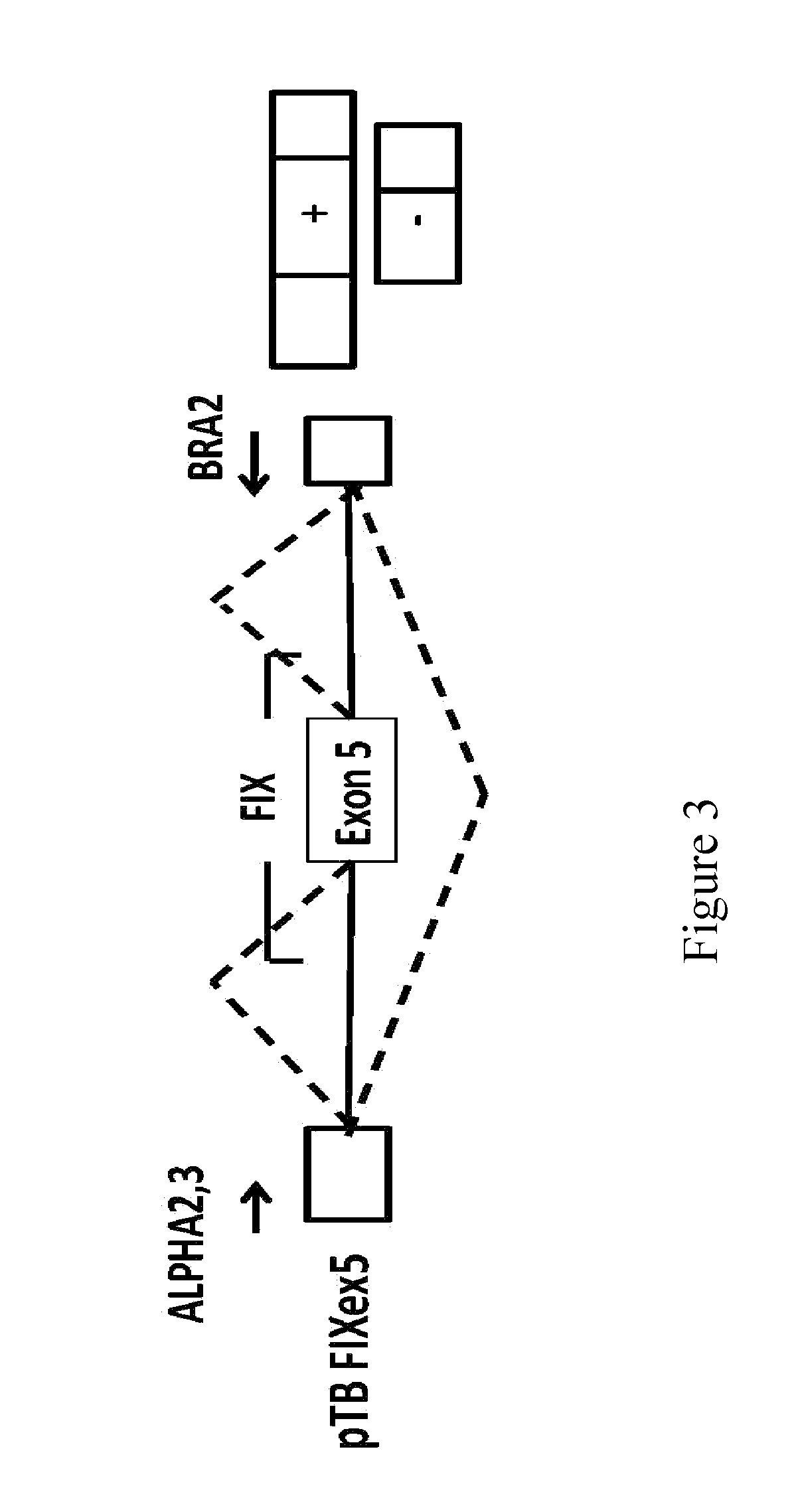
tations Near the Donor Site and Mutations in the Poly-Pyrimidine Sequence Upstream of the Exon 5 Acceptor Site of the Coagulation Factor IX Associated with Hemophilia B
[0078]In the factor IX gene (F9), the exonic mutations at position −2 within the donor site, as well as the mutations at positions −8 and −9 within the acceptor site of exon 5, are associated with hemophilia B. It is interesting to note that the mutations at position −2 in the exon are synonymous and do not modify the coding sequence but induce exon skipping and therefore they are classifiable as splicing mutations. The mutations at positions −8 and -9 within the acceptor site also induce skipping of exon 5.
[0079]Table 2 shows the mutations under discussion which were identified in patients affected by hemophilia B (Hemophilia B International database). Nucleotides belonging to exon 5 are shown in capital letters, whereas those belonging to the intron are in lower case. Each position, shown at the bottom of the figure...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Composition | aaaaa | aaaaa |
| Therapeutic | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com