Cas9 effector-mediated regulation of transcription, differentiation and gene editing/labeling
a technology of transcription factor and effector, applied in the field of dcas9-effector system, can solve the problems of inability to influence the differentiation status of stem cells, and the inability of dcas9-effector system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cas9 Effector-Mediated Regulation of Transcription and Differentiation in Human Pluripotent Stem Cells
[0170]a. sgRNA in Silico Design
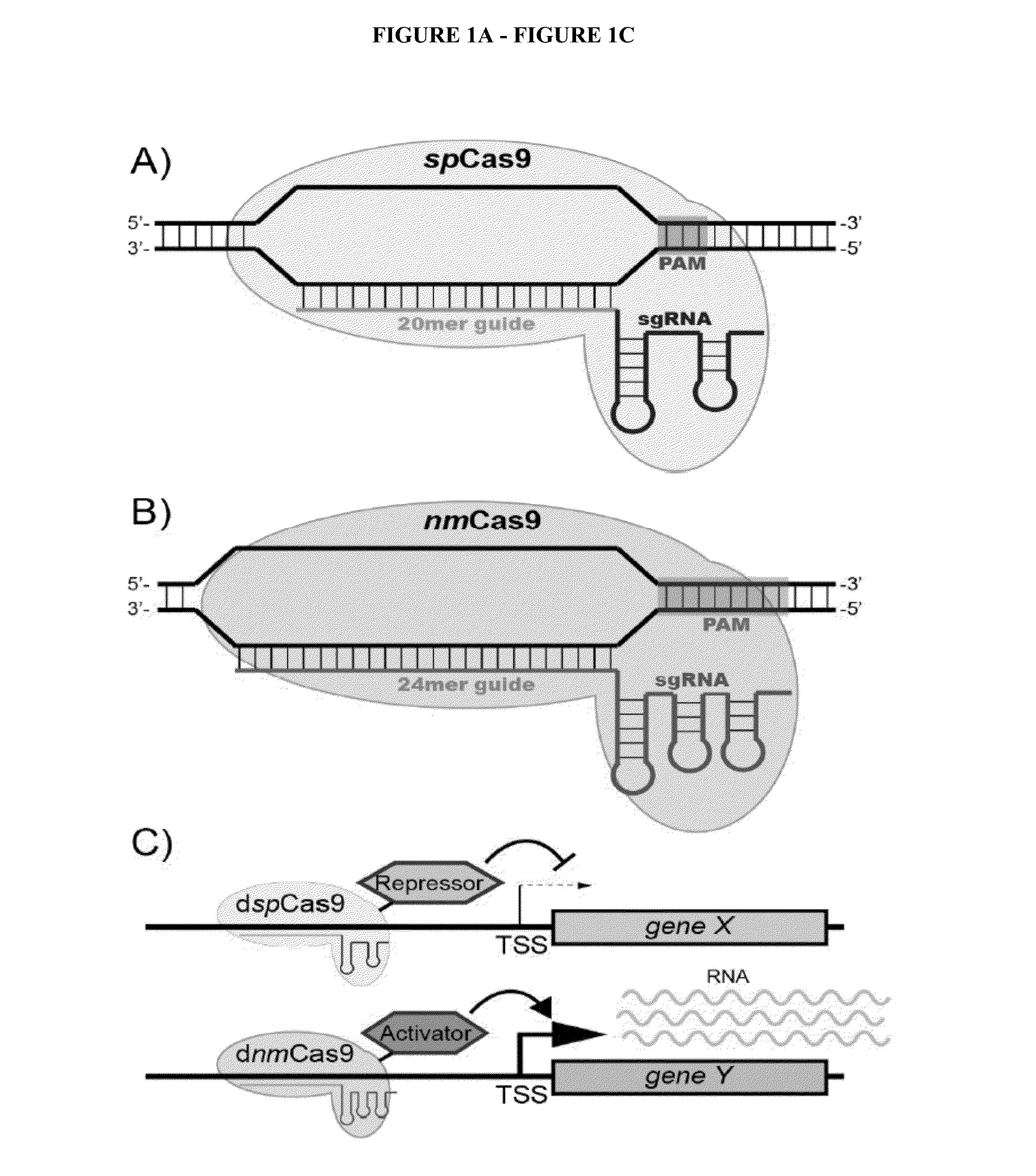
[0171]Candidate sgRNAs were identified by searching for (G(N)20GG) motifs 300 bases upstream of the and 100 bases downstream of the transcriptional start site (TSS) that conform with the nucleotide requirements for U6 Pol III transcription and the spCas9 PAM recognition element (NGG) (Jinek et al., 2012 [23]; Mali et al., 2013b [4]). Bowtie2 was used to map candidate targets to the human genome (build GRCh37) (Langmead and Salzberg, 2012 [54]) with sensitive parameters (--local-f-k10--very-sensitive-local-L9-N1) to detect potential off-target sites. All the sgRNAs used herein had no other genomic matches at the alignment stringency used. See, Table 3.
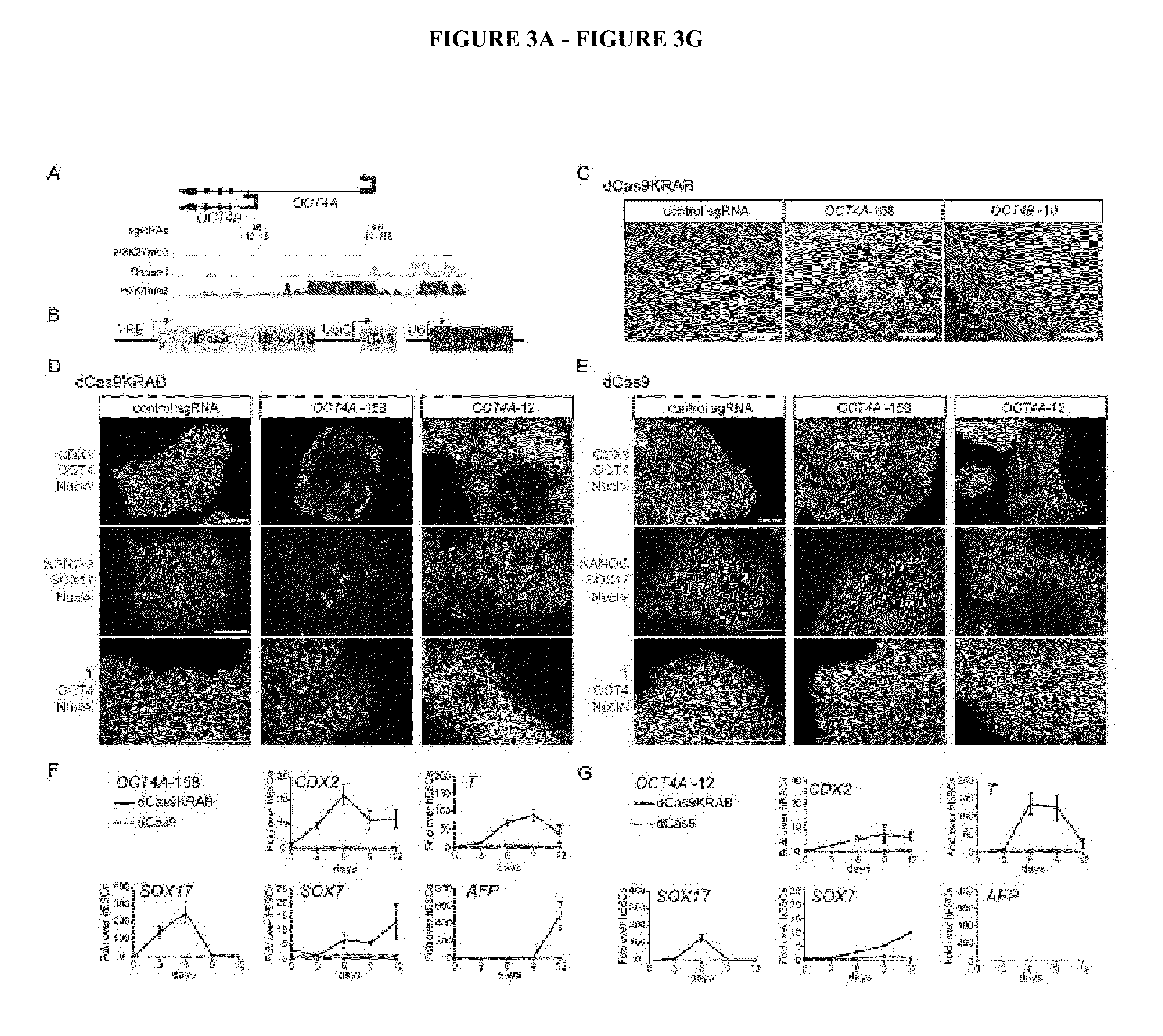
TABLE 3sgRNAsPosi-TargettionPro-toTargetTarget SequenceNamemoterTSSStrand(including PAM)OCT4Aoct4−158templateGGGGCGCCAGTTGTGTCTCCCGGisoform(SEQ ID No: 1)AOCT4Aoct4−12templateGTGGGACTGGGGAGGGAGAGAGGisoform(...
example 2
dCas9-Mediated Reprogramming of Human Fibroblasts to iPSCs
[0186]Since the groundbreaking work by Yamanaka and colleagues [15] that demonstrated the feasibility of reprogramming cellular identity with OCT4, SOX2, KLF4 and cMYC (OSKM), intense scientific effort has focused on understanding the mechanism of this process and improving it through the identification of additional collaborating TFs and the substitution / inclusion of small molecules or non-coding RNAs [25]. Artificial TFs that activate expression of individual TFs within the OSKM set can substitute for a single factor (e.g. SKM with a TALE-VP64 fusion that activates OCT4 can reprogram fibroblasts to iPSCs [36]). Fibroblast reprogramming to iPSCs will be used as a framework for the initial demonstration the multi-target activation via dspCRSIPRa can yield functional differentiation state choices. Initial experiments will focus on the iterative substitution of single OSKM factor with a dspCas9-VP64 effector targeting one of th...
example 3
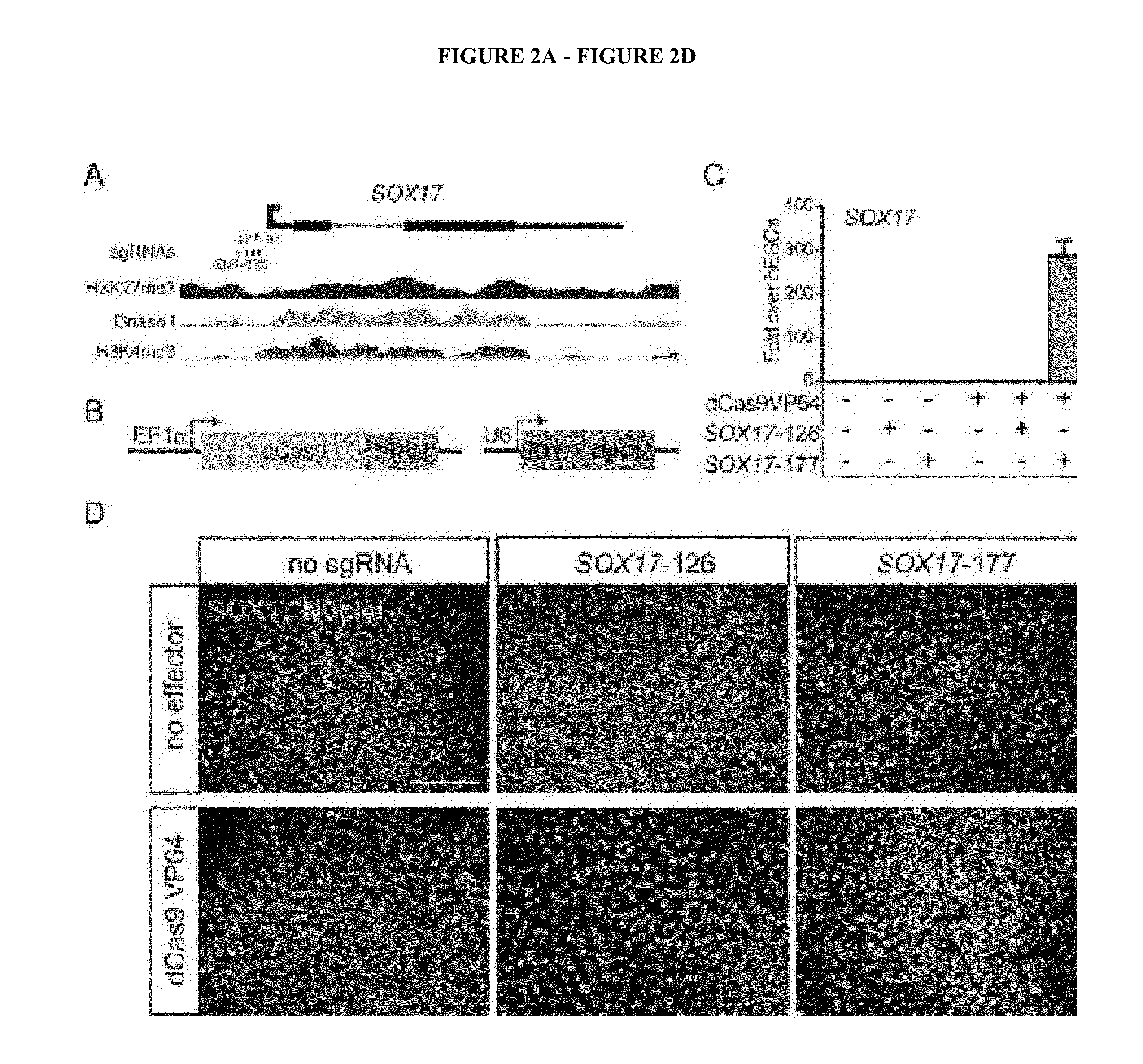
[0188]Identification Of Factors Generating A Definitive Endoderm (DE) From hESCs
[0189]The first major differentiation state from ESCs to endodermal lineages may involve the transition to DE [60-62]. Monolayer cell culture conditions that efficiently generate DE through activation of the wingless (WNT) and TGFβ signaling pathways are well defined [60, 63]. This well-defined lineage will be used to test the ability of the CRISPRe / i system to program cell fate decisions (Schematic overview of the approach given in FIG. 8A). As a testing ground, directing hESCs to an endodermal fate has several benefits: First, diagnostic markers of DE have been defined (e.g. CXCR4, GATA3, FOXA3, etc.) [60, 63-66], and detection of intracellular SOX17 with FOXA2 or surface expression of CXCR4 and c-Kit have been used to distinguish DE from ESCs by FACS [60, 64, 67]. This provides a robust and sensitive assay for the identification of cells that have differentiated from hESCs to DE. Second, critical fact...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Distance | aaaaa | aaaaa |
| Fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com