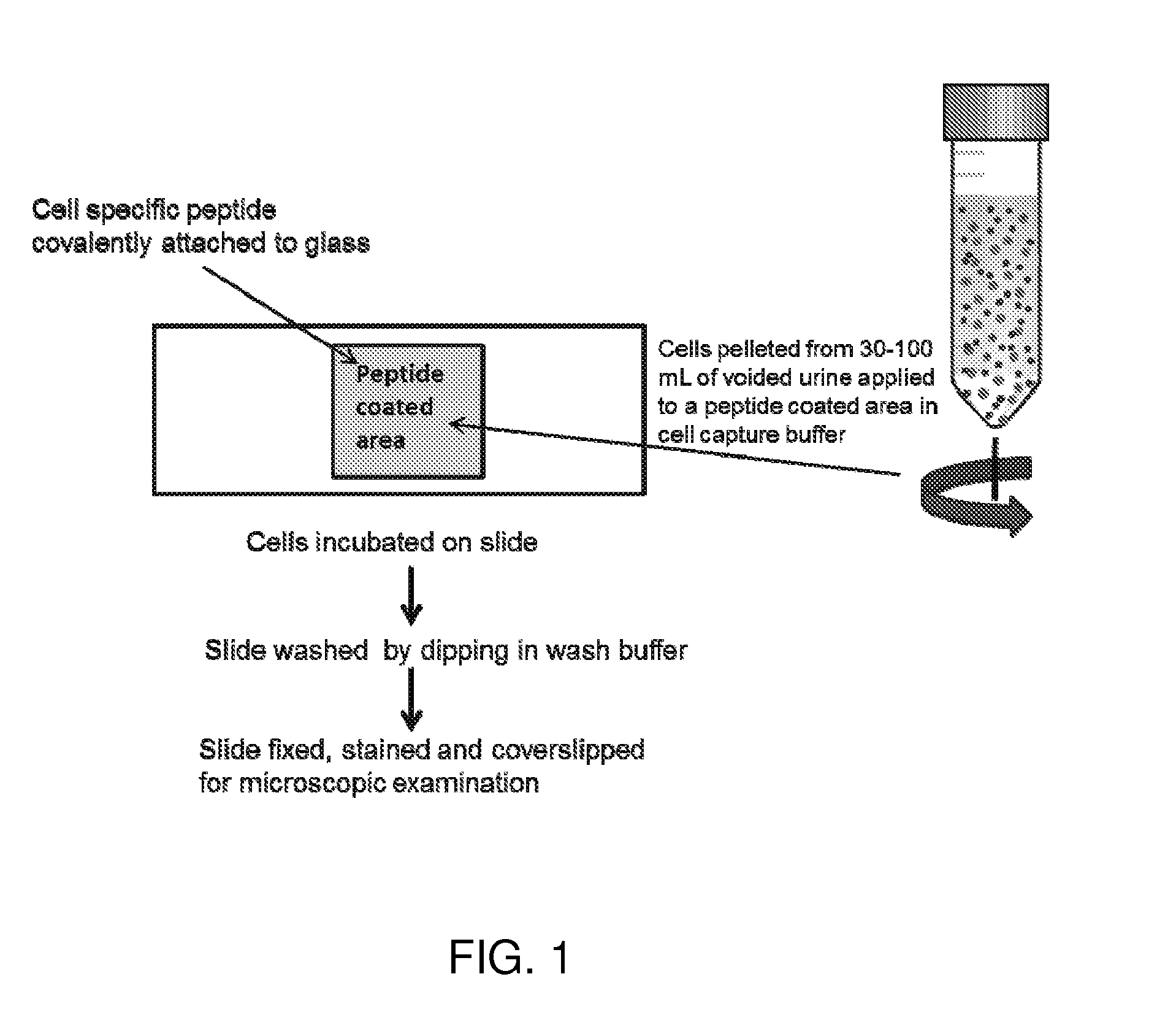
Cell binding peptides for diagnosis and detection
a technology of cell binding and detection, applied in the direction of peptides, peptide/protein ingredients, instruments, etc., can solve the problems of cytology and slide-based diagnostic testing being confounded, putting a significant financial burden on both the patient and the healthcare facility, and requiring expensive methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Generation of Synthetic Binding Peptides
[0063]Peptide Synthesis:
[0064]Binding peptide sequences were synthesized using standard Solid-Phase peptide synthesis techniques on a SYMPHONY Peptide Synthesizer (Protein Technologies, Inc., Tucson, Ariz.) using standard Fmoc / t-Bu chemistry with the following coupling reagents at a 1:1:1:2 ratio—Amino acids; O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluoroborate (HBTU); 1-hydroxybenzotriazole (HOBt); and N-Methylmorpholine (NMM). A20% (v / v) solution of piperidine in DMF was utilized for Fmoc removal. Peptides were synthesized on Fmoc-NH-Rink-Ahx-MBHA resin or on Fmoc-PAL-Peg-PS resin yielding peptides with C-terminal amides.
[0065]Amino acids with orthogonal side-chain protecting groups were coupled in 5-fold excess in the synthesis cycles, and all residues were doubly or triply coupled for 1 h. The coupling reactions were monitored by Kaiser ninhydrin test or chloranil test. After all amino acid residues were coupled and followin...
example 2
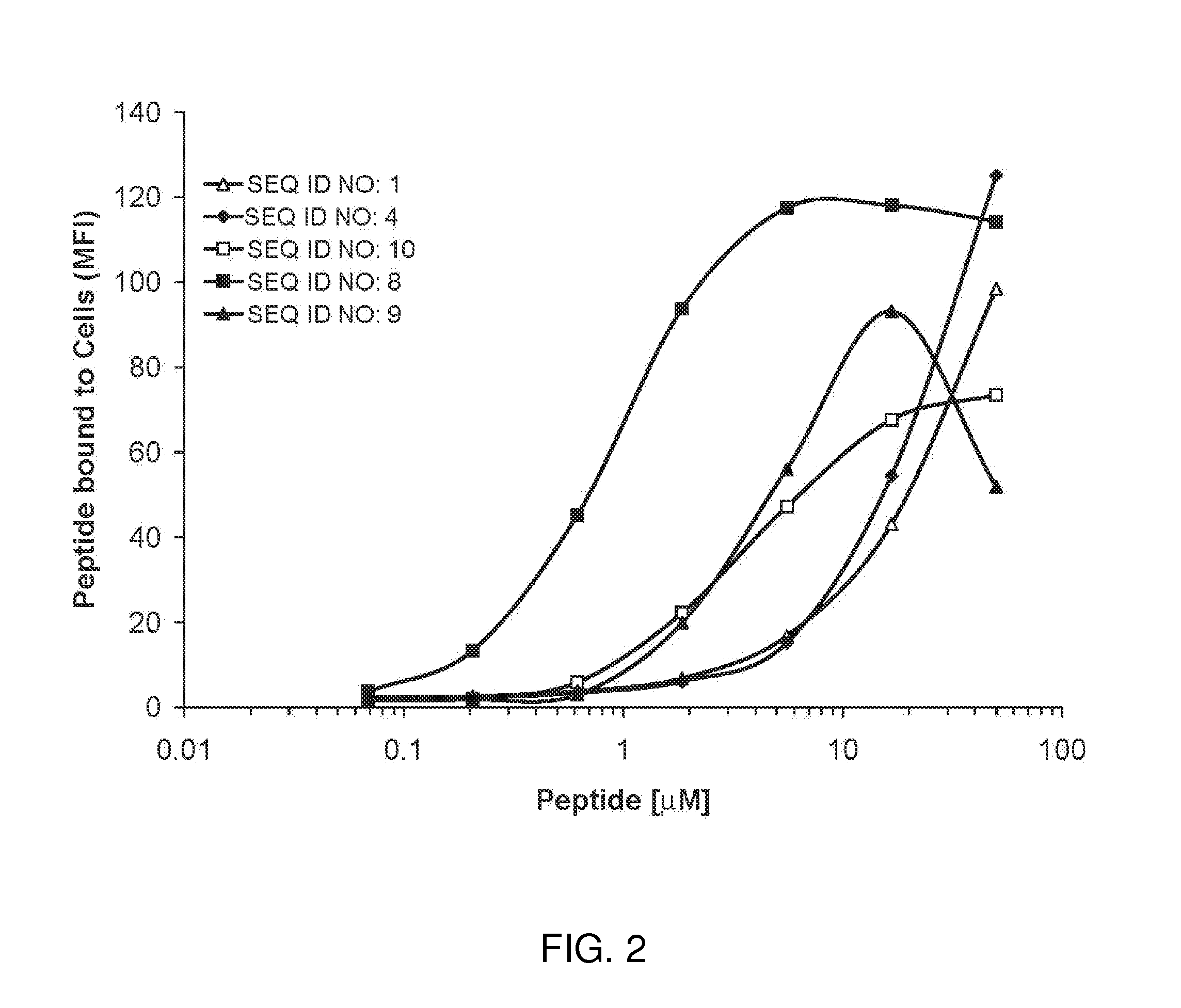
[0066]Synthetic biotinylated cell binding peptides were examined for their ability to bind to a bladder cancer cell line, J82 (ATCC, HTB1). The cell binding peptide was synthesized with a PEG-10 spacer and a carboxyl terminal lysine-biotin. J82 cells were grown to confluency on tissue-culture treated polystyrene. Cells were washed with PBS and released from the support with 0.05% trypsin. Trypsin was removed by centrifuging cells at 500×g and resuspending in PBS supplemented with 0.5% BSA and 2 mM EDTA to a concentration of 68,000 cells / ml. Aliquots of cells (50 μL) were separately incubated in 100 μl PBS / 0.5% BSA / 2 mM EDTA containing a final concentration of 0, 0.07, 0.21, 0.62, 1.85, 5.55, 16.7, and 50 μM peptide for 60 min at 4° C. Cells were then washed thrice in 1.8 ml PBS / 0.5% BSA / 2 mM EDTA with 500×g centrifugation for 5 min between washes. Approximately 50 μl of wash buffer remained with the cells after the final wash. Fluorescently-tagged neutravidin (Ne...
example 3
Capture of Cultured J82 Tumor Cells from Urine with Cell Binding Peptide Attached to a Support
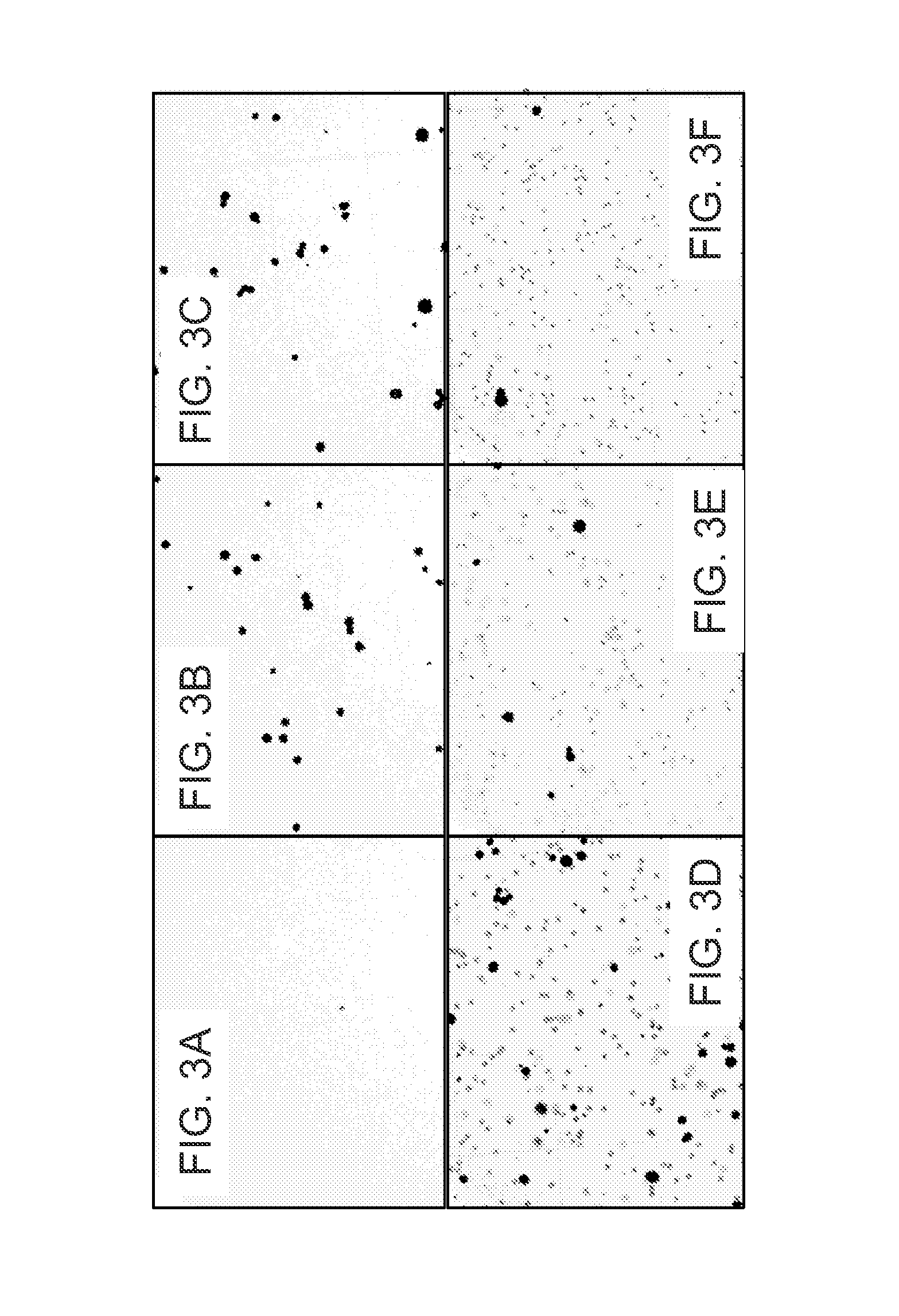
[0067]To demonstrate the ability of cell binding peptides to recover tumor cells from urine, biotinylated peptides were immobilized on Streptavidin coated magnetic beads and used to separate tumor cells from white blood cells.
[0068]A green fluorescent leukocyte preparation was prepared as follows. Red blood cells in 2 ml anti-coagulated human blood were lysed by the addition of 30 ml of a hypotonic medium and incubation for 10 min at room temperature. Intact cells and debris were recovered by centrifugation at 300×g. Pelleted material was resuspended with PBS / 0.5% BSA / 2 mM EDTA and filtered through a 100 micron nylon filter. Leukocytes (3×106) were labeled with 5 μM Cell Tracker Green CMFDA (LIFE TECHNOLOGIES / MOLECULAR PROBES) in EMEM growth medium without serum for 45 min. Excess dye was removed by centrifugation at 500×g for 5 min and washing with PBS / 0.5% BSA / 2 mM EDTA and cells were exp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| RH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| fluorescence in- | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com