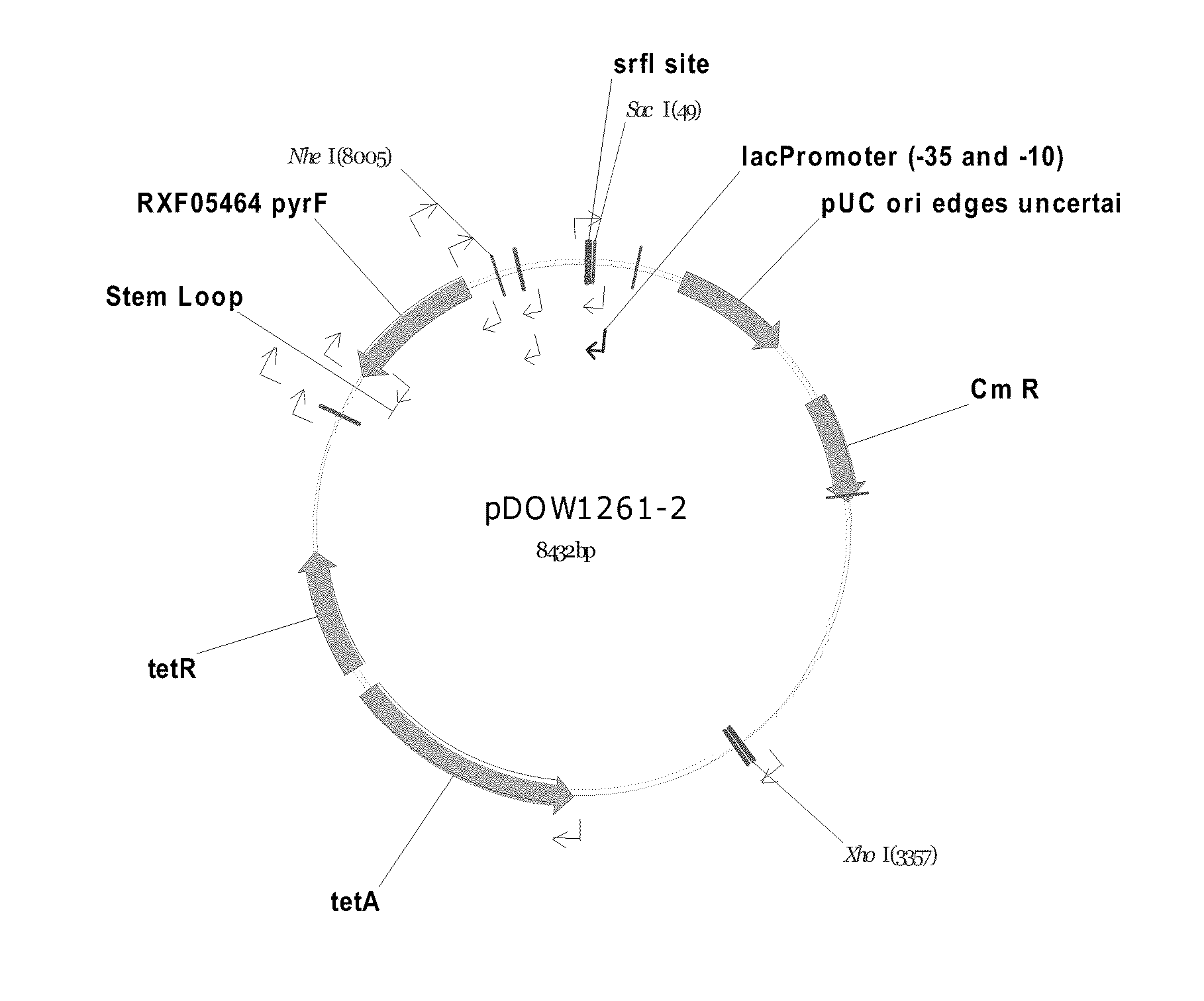
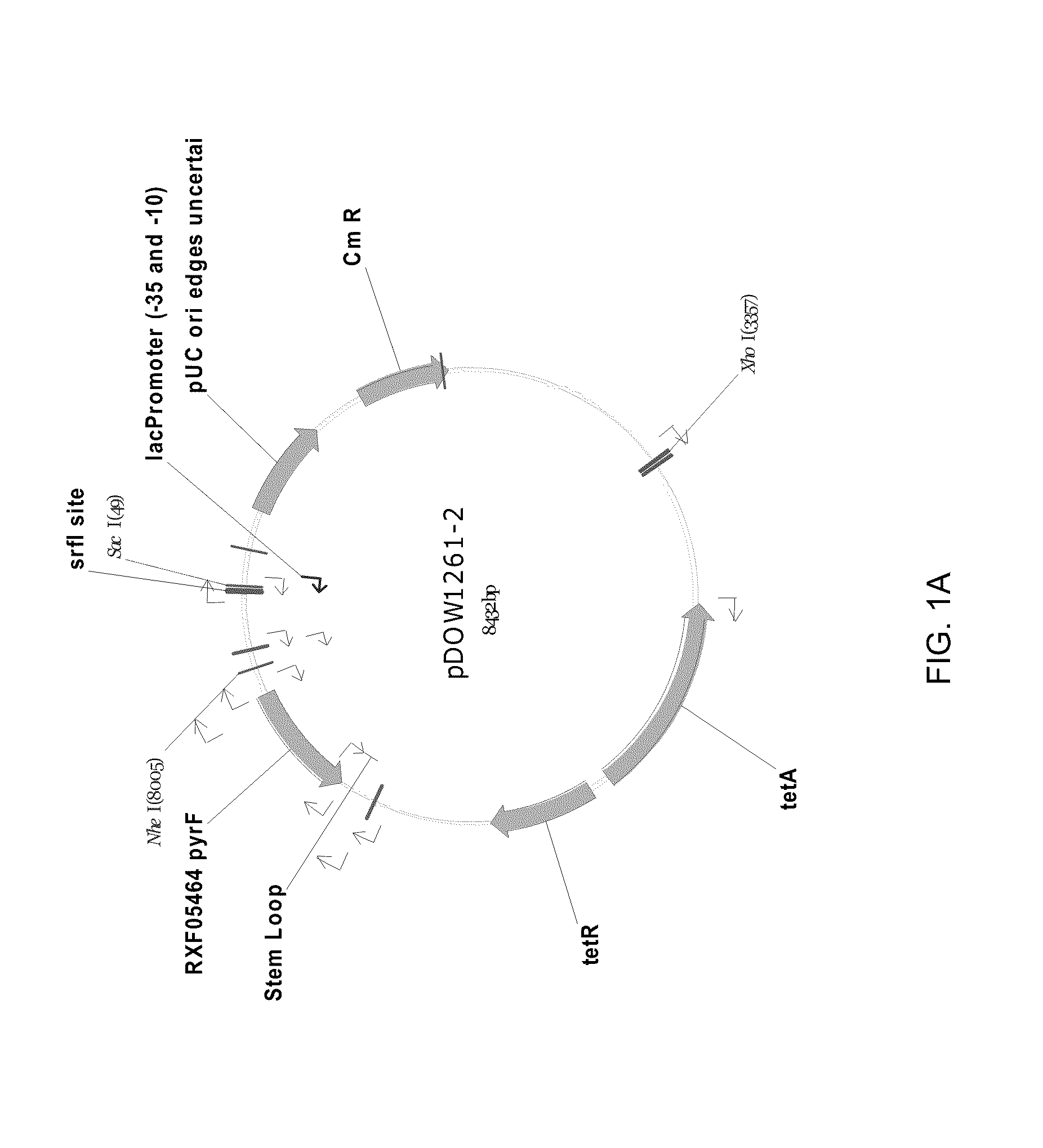
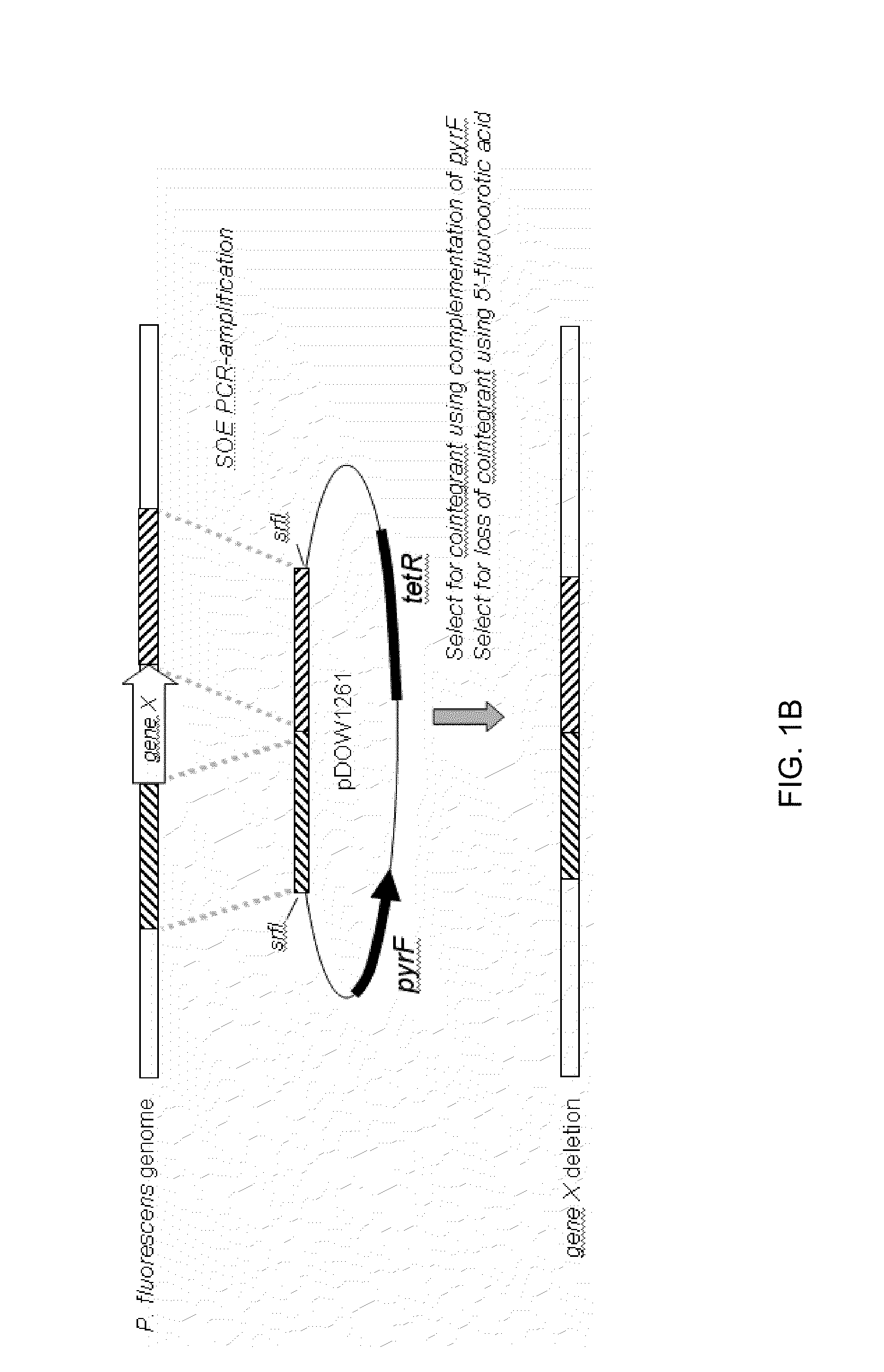
Method for Rapidly Screening Microbial Hosts to Identify Certain Strains with Improved Yield and/or Quality in the Expression of Heterologous Proteins
a technology of heterologous protein and microbial host, which is applied in the field of protein production, can solve the problems of inefficient secondary modification, insoluble or improperly folded proteins, and inability to rapidly detect and identify heterologous protein expression strains, so as to improve yield and/or quality, increase and reduce the expression of one or more target genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
experimental examples
Overview
[0232]Heterologous protein production often leads to the formation of insoluble or improperly folded proteins, which are difficult to recover and may be inactive. Furthermore, the presence of specific host cell proteases may degrade the protein of interest and thus reduce the final yield. There is no single factor that will improve the production of all heterologous proteins. Thus, a method was sought to identify factors specific to a particular heterologous protein from a pool of likely candidates.
[0233]Using Systems Biology tools, the P. fluorescens genome was mined to identify host cell protein folding modulator and protease genes. Then, global gene expression analyses were performed to prioritize upregulated targets, and, thereafter, novel protein production strains were constructed. As a result, a “Pfenex Strain Array” was assembled consisting of a plurality of phenotypically distinct P. fluorescens host strains that are deficient in host-cell proteases or allow the co-...
example 1
Identification of Folding Modulator Genes in the Genome of P. fluorescens Strain MB214
[0235]Folding modulators are a class of proteins present in all cells which aid in the folding, unfolding and degradation of nascent and heterologous polypeptides. Folding modulators include chaperones, chaperonins, peptidyl-prolyl cis-trans isomerases, and proteins involved in protein disulfide bond formation. As a first step to construct novel production strains with the ability to help fold heterologous proteins, the P. fluorescens genome was mined to identify host cell folding modulator genes.
[0236]Each of the 6,433 predicted ORFs of the P. fluorescens MB214 genome was analyzed for the possibility that they encoded a folding modulator using the following method. Several folding modulators of interest had already been identified by Dow researchers by analysis of the genome annotation (Ramseier et. al. 2001). Homologs of these starting proteins were identified using protein / protein BLAST with the...
example 2
Identification of Protease Genes in the Genome of P. fluorescens Strain MB214
[0240]Proteases are enzymes that hydrolyze peptide bonds and are necessary for the survival of all living creatures. However, their role in the cell means that proteases can be detrimental to recombinant protein yield and / or quality in any heterologous protein expression system, which also includes the Pfenex Expression Technology™. As a first step to construct novel production strains that have protease genes removed from the genome, the P. fluorescens genome was mined to identify host cell protease genes.
[0241]Each of the 6,433 predicted ORFs of the P. fluorescens MB214 genome were analyzed for the possibility that they encoded a protease using the following method. The MEROPS database is manually curated by researchers at the Wellcome Trust Sanger Institute, Cambridge, UK (Rawlings et. al. 2006, http: / / merops.sanger.ac.uk). It is a comprehensive list of proteases discovered both through laboratory experi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com