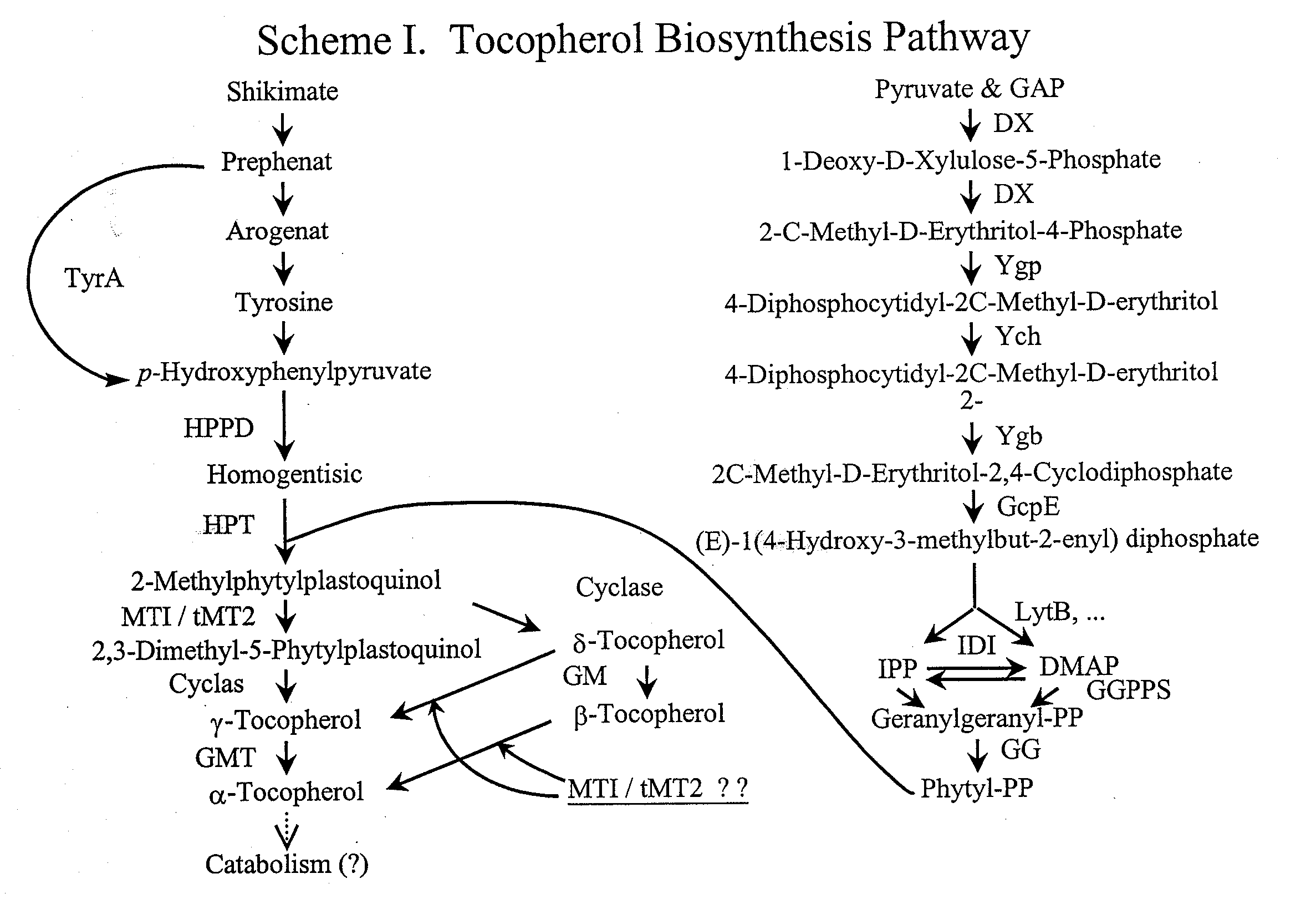
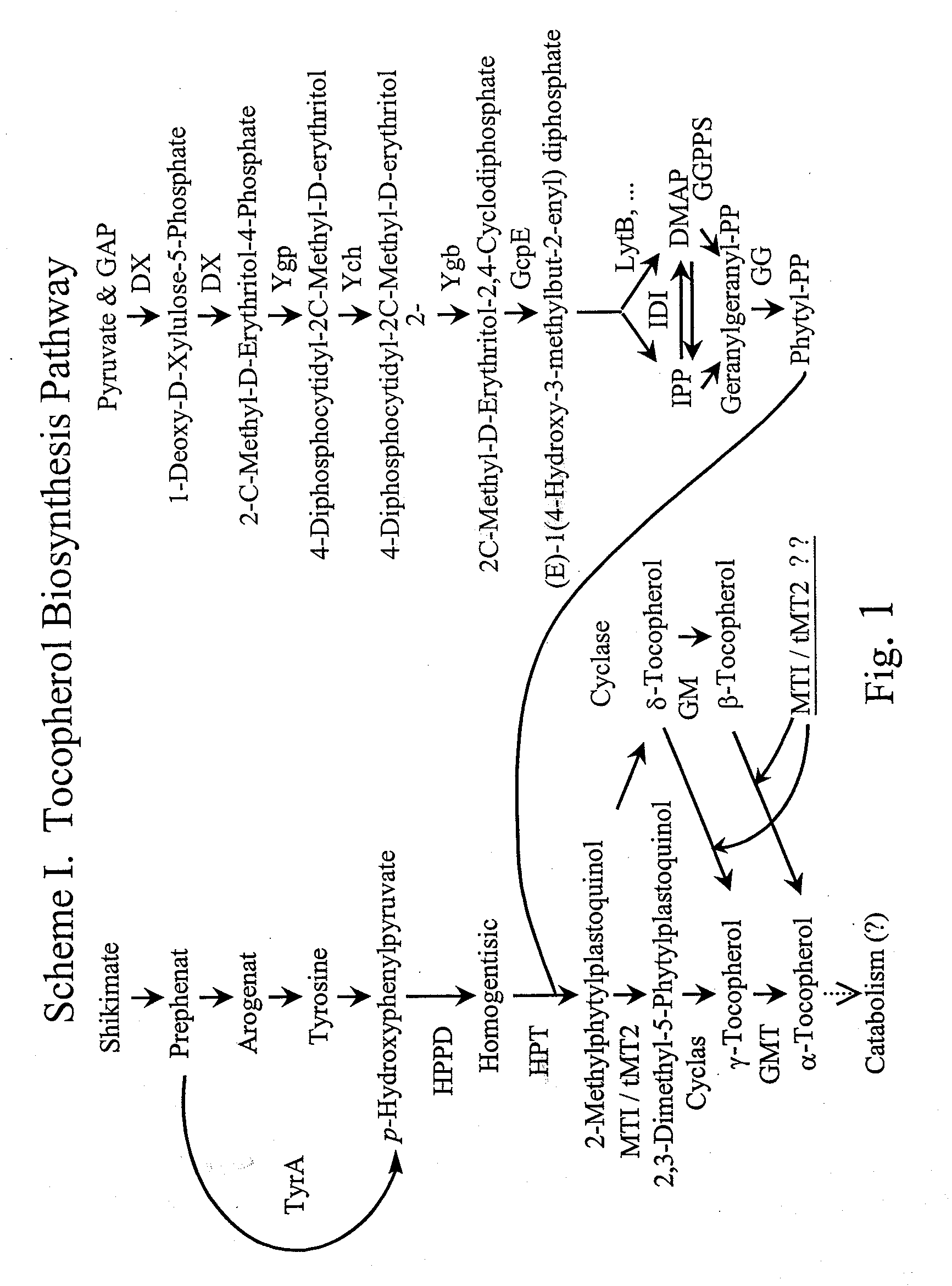
Aromatic methyltransferases and uses thereof
a technology of methyltransferases and methyltransferases, which is applied in the direction of transferases, enzymology, gymnosperms, etc., can solve the problems of relatively high supplementation costs and difficult to achieve the recommended daily intake of 15-30 mg of vitamin e from the average american diet, so as to and increase the -tocopherol level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification and Characterization of Mutant hdt2 Arabidopsis thaliana, Ecotype Landsberg Plants
[0378] Mutagenized (M2) seeds of Arabidopsis thaliana, ecotype Landsberg are obtained both by purchase from Lehle Seeds (Round Rock, Tex., U.S.A.) and by standard EMS mutagenesis methodology. The M2 plants are grown from the M2 seeds in greenhouse conditions with one plant per 2.5 inch pot. The resulting M3 seeds are collected from individual M2 plants and analyzed for tocopherol levels.
[0379] Seeds from approximately 10,000 M3 lines of Arabidopsis thaliana, ecotype Landsberg or Col-O are analyzed for individual tocopherol levels using the following procedure. Five milligrams of seeds from individual plants are ground to a fine powder using a ⅛″ steel ball bearing and vigorous shaking. 200 Microliters of 99.5% ethanol / 0.5% pyrogallol is added, mixed for 30 seconds and allowed to incubate at 4° C. for 1 h. 50 Microgram / ml of tocol (Matreya, Inc., Pleasant Gap, Pa.) is added to each samp...
example 2
Identification and Sequencing of the Mutant hdt2 Gene in the Arabidopsis thaliana, Landsberg Erecta (Ler) High δ-Tocopherol Mutants
[0381] Using map-based cloning techniques (see, for example, U.S. Ser. No. 09 / 803,736, Plant Polymorphic Markers and Uses Thereof, filed Mar. 12, 2001) the mutant hdt2 gene is mapped to chromosome 3 telomeric marker T12C14—1563 at 85 cM. This region contains approximately 60 predicted genes. Our analysis of the genes in this region revealed that one of the genes, MAA21—40, possesses homology to known ubiquinone methyltransferases. Based on this homology and the prediction that MAA21—40 is targeted to the chloroplast, this gene is determined to be likely to contain the mutation responsible for the high δ-tocopherol phenotype in hdt2 mutants. The sequences of the MAA21—40 gene locus in the wild types and hdt2 mutants are PCR amplified, and determined by standard sequencing methodology. The gene locus, in each case, is amplified using the sequencing primer...
example 3
Identification of Genes from Various Sources Demonstrating Homology to the tMT2 Gene from Arabidopsis thaliana
[0398] The protein sequence of tMT2 from Arabidopsis thaliana (NCBI General Identifier Number gi7573324) is used to search databases for plant sequences with homology to tMT2 using TBLASTN (Altschul et al., Nucleic Acids Res. 25:3389-3402 (1997); see also www.ncbi.nlm.nih.gov / BLAST / ). Nucleic acid sequences SEQ ID NO: 8 through 15 are found to have high homology with the Arabidopsis sequence.
>CPR19219 Brassica napus tMT2 homolog 1 - LIB4153-013-R1-K1-B7(SEQ ID NO: 13)ATGGCTTCTCTCATGCTCAACGGGGCCATCACCTTCCCCAAGGGATTAGGCTTCCCCGCTTCCAATCTACACGCCAGACCAAGTCCTCCGCTGAGTCTCGTCTCAAACACAGCCACGCGGAGACTCTCCGTGGCGACAAGATGCAGCAGCAGCAGCAGCGTGTCGGCGTCAAGGCCATCTGCGCAGCCTAGGTTCATCCAGCACAAGAAAGAGGCCTACTGGTTCTACAGGTTCCTGTCCATCGTGTACGACCACATCATCAATCCCGGCCACTGGACGGAGGATATGAGGGACGACGCTCTCGAGCCTGCGGATCTGAGCCATCCGGACATGCGAGTTGTCGACGTCGGAGGCGGAACGGGTTTCACCACGCTGGGAATCGTCAAGACGGTGAAGGCTAAGAACGTGACGA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| weight | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com