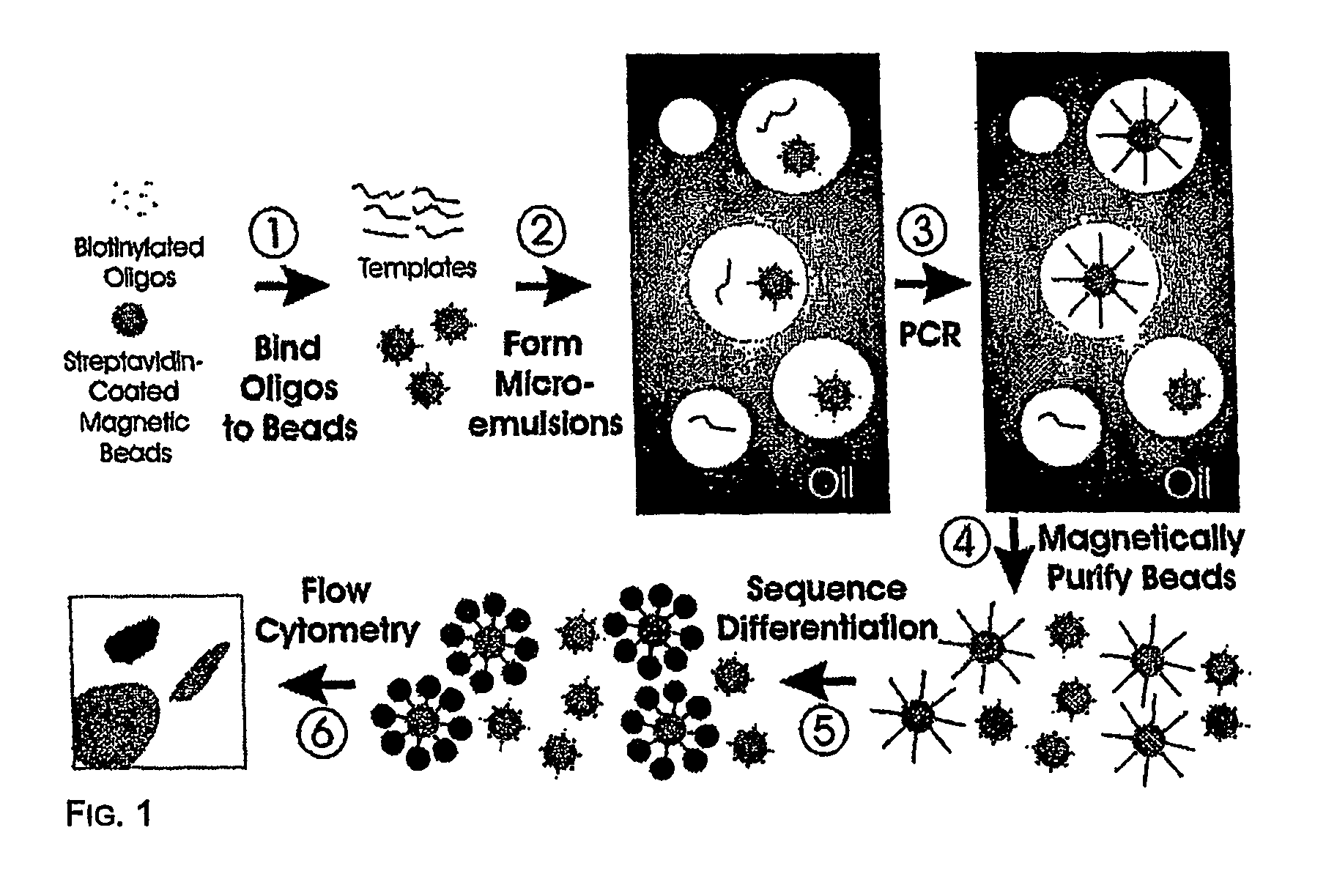
Method and compositions for detection and enumeration of genetic variations
a technology of genetic variation and composition, applied in the field of genetic analysis, can solve the problems of complex analysis, limited digital technology to counting tens to thousands of molecules, and limited accuracy and sensitivity of instruments and experimental nois
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
[0045] Step 1—Coupling oligonucleotides to beads. Superparamagnetic beads of 1.05+ / −0.1 um in diameter, covalently bound to streptavidin, were purchased from Dynal Biotech, Inc. (650.01, Lake Success, N.Y.). Beads were washed once with 1×PCR buffer (53286, Invitrogen, Carlsbad, Calif.) then suspended in Bind and Wash Buffer (BWB) (5 mM Tris-HCl, 0.5 mM EDTA, 1.0 M NaCl, pH 7.5). Beads were incubated in BWB for 30 min at room temperature in the presence of 10 uM oligonucleotides (FIG. 8). These oligonucleotides were modified with a dual biotin group at the 5′ end with the biotin groups separated by a six-carbon linker (IDT, Coralville, Iowa). After binding, the beads were washed 3 times with 1×PCR buffer to thoroughly remove unbound oligonucleotides.
[0046] Step 2—Preparing microemulsions. Microemulsions for PCR were prepared by slight modifications of previously described methods (14) (15). The oil phase was composed of 4.5% Span 80 (S6760, Sigma, St. Louis, M...
example 2
Results
[0053] Step 1—Coupling oligonucleotides to beads. We used streptavidin-beads because of the simplicity of coupling biotinylated oligonucleotides to them. Oligonucleotides with just a single 5′ biotin group were found to dissociate from the beads during temperature cycling, while oligonucleotides labeled with dual biotin groups at their 5′ end (separated by a six-carbon linker) were stable to cycling. As determined by fluoroscopic measurements of oligonucleotides doubly labeled with 6-FAM and biotin, ˜105 oligonucleotide molecules were bound to each bead. We found that short oligonucleotides (20 bases) did not work as well for priming as longer ones (41 bp), perhaps because of steric hindrance at the bead surface. It is likely that amino-, sulfhydryl-, or carboxyl-modified oligonucleotides covalently coupled to beads modified with corresponding reactive groups could also function as bead-bound primers for BEAMing.
[0054] Step 2—Preparing microemulsions. The size of the indiv...
example 3
Characteristics of Microemulsions.
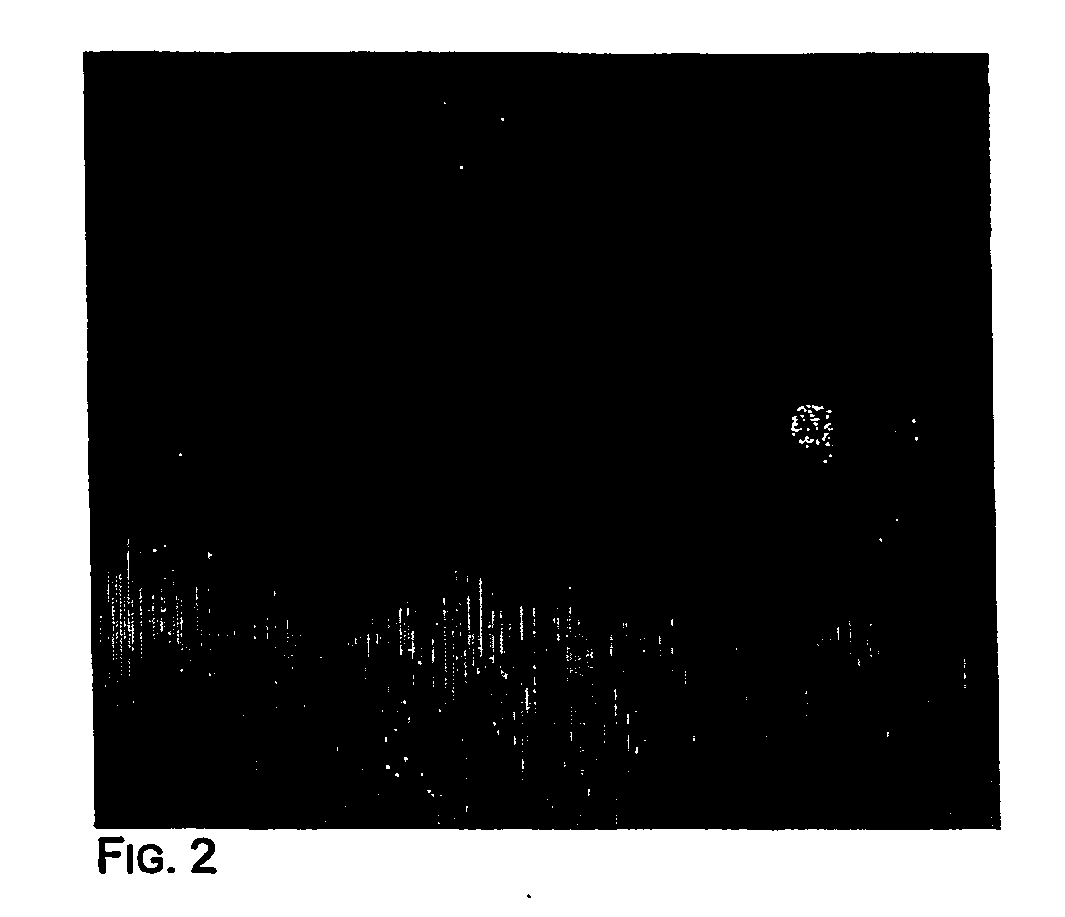
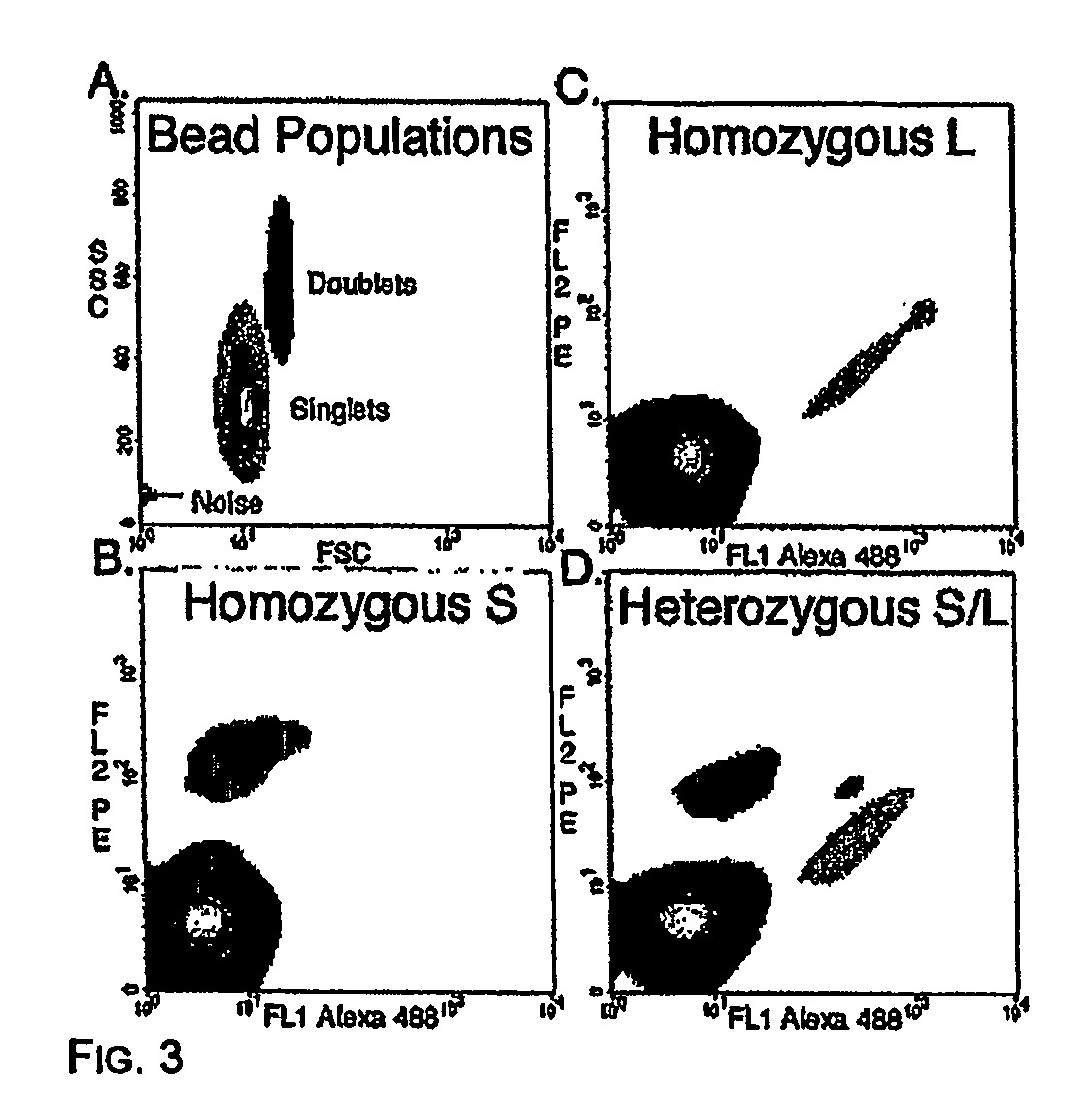
[0059] Pilot experiments demonstrated that simply stirring the water-oil mixtures described in Materials and Methods produced very stable microemulsions of a size compatible with that of the beads. In the experiment shown in FIG. 2, the aqueous compartment contained a blue dye and 1 micron magnetic beads that were labeled by binding to an oligonucleotide that was biotinylated at its 5′ end and labeled with fluorescein at its' 3′ end The appearance of emulsions immediately after their formation is shown in FIG. 2. As expected, this appearance was unchanged after temperature cycling during PCR (15). Most aqueous compartments contained no beads, as expected from the figures provided in the previous section. Those compartments that did contain beads generally contained only one, though a fraction contained more, as expected from a Poisson distribution and non-uniform aqueous compartment sizes. “Heterozygous” beads containing PCR products representing ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com